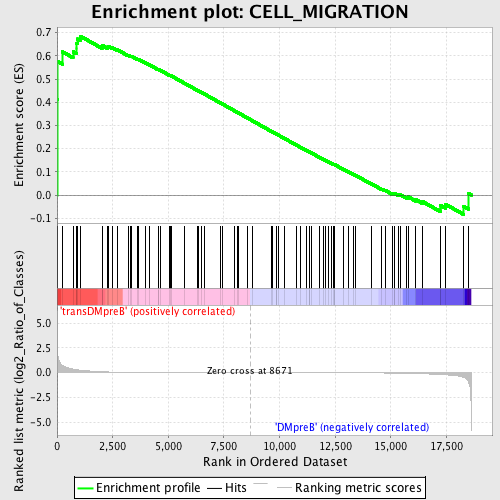
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | CELL_MIGRATION |
| Enrichment Score (ES) | 0.6852777 |
| Normalized Enrichment Score (NES) | 1.5189474 |
| Nominal p-value | 0.007434944 |
| FDR q-value | 0.4518405 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MYH9 | 2252 2244 | 1 | 5.596 | 0.4118 | Yes | ||
| 2 | IL10 | 14145 1510 1553 22902 | 14 | 2.242 | 0.5762 | Yes | ||
| 3 | PPAP2B | 7503 16161 | 238 | 0.729 | 0.6178 | Yes | ||
| 4 | DOCK2 | 8243 | 718 | 0.354 | 0.6181 | Yes | ||
| 5 | IL12A | 4913 | 864 | 0.303 | 0.6326 | Yes | ||
| 6 | ABI3 | 7314 | 865 | 0.303 | 0.6548 | Yes | ||
| 7 | SYK | 21636 | 921 | 0.288 | 0.6730 | Yes | ||
| 8 | VCL | 22083 | 1039 | 0.252 | 0.6853 | Yes | ||
| 9 | THY1 | 19481 | 2023 | 0.090 | 0.6389 | No | ||
| 10 | ITGB2 | 19978 | 2033 | 0.089 | 0.6449 | No | ||
| 11 | LAMC1 | 13805 | 2258 | 0.071 | 0.6380 | No | ||
| 12 | MDGA1 | 13231 | 2312 | 0.067 | 0.6401 | No | ||
| 13 | PLG | 1593 23383 | 2488 | 0.057 | 0.6349 | No | ||
| 14 | SLIT1 | 23678 | 2717 | 0.046 | 0.6260 | No | ||
| 15 | RTN4 | 7569 1328 1343 1442 | 3201 | 0.032 | 0.6022 | No | ||
| 16 | NF1 | 5165 | 3307 | 0.029 | 0.5987 | No | ||
| 17 | WNT1 | 22371 | 3341 | 0.028 | 0.5990 | No | ||
| 18 | PPAP2A | 21562 3258 3244 | 3625 | 0.023 | 0.5854 | No | ||
| 19 | PF4 | 16800 | 3668 | 0.022 | 0.5848 | No | ||
| 20 | NF2 | 1222 5166 | 3971 | 0.018 | 0.5699 | No | ||
| 21 | UNC5C | 10256 | 4166 | 0.016 | 0.5606 | No | ||
| 22 | NRD1 | 16140 | 4546 | 0.013 | 0.5411 | No | ||
| 23 | PARD6B | 12209 | 4569 | 0.013 | 0.5409 | No | ||
| 24 | CD2AP | 22975 | 4626 | 0.013 | 0.5388 | No | ||
| 25 | SAA1 | 9776 | 5069 | 0.010 | 0.5157 | No | ||
| 26 | SEMA3B | 5422 | 5074 | 0.010 | 0.5162 | No | ||
| 27 | CX3CL1 | 9791 | 5134 | 0.010 | 0.5137 | No | ||
| 28 | MIA3 | 6868 6867 11679 | 5714 | 0.007 | 0.4830 | No | ||
| 29 | CALCA | 4470 | 6322 | 0.005 | 0.4507 | No | ||
| 30 | CD34 | 14006 | 6354 | 0.005 | 0.4494 | No | ||
| 31 | SLIT2 | 5456 | 6511 | 0.005 | 0.4413 | No | ||
| 32 | CNTN4 | 11265 1088 17345 | 6610 | 0.005 | 0.4364 | No | ||
| 33 | IL12B | 20918 | 7351 | 0.003 | 0.3966 | No | ||
| 34 | ACVRL1 | 22354 | 7452 | 0.003 | 0.3914 | No | ||
| 35 | OTX2 | 9519 | 7990 | 0.001 | 0.3626 | No | ||
| 36 | ITGB1BP1 | 9189 | 8107 | 0.001 | 0.3564 | No | ||
| 37 | ANGPTL3 | 16171 | 8143 | 0.001 | 0.3546 | No | ||
| 38 | ALOX15B | 20397 | 8571 | 0.000 | 0.3315 | No | ||
| 39 | NRP2 | 5194 9486 5195 | 8763 | -0.000 | 0.3213 | No | ||
| 40 | SHH | 16581 | 9614 | -0.002 | 0.2756 | No | ||
| 41 | NRXN3 | 2153 5196 | 9670 | -0.002 | 0.2728 | No | ||
| 42 | ABI2 | 4037 6830 | 9876 | -0.003 | 0.2619 | No | ||
| 43 | TNN | 11625 | 9881 | -0.003 | 0.2619 | No | ||
| 44 | NRXN1 | 1581 1575 22875 | 9958 | -0.003 | 0.2580 | No | ||
| 45 | GDNF | 22514 2275 | 10216 | -0.003 | 0.2444 | No | ||
| 46 | AMOT | 11341 24037 6587 | 10754 | -0.005 | 0.2158 | No | ||
| 47 | SEMA4F | 17105 | 10959 | -0.005 | 0.2051 | No | ||
| 48 | THBS4 | 21393 | 11201 | -0.006 | 0.1926 | No | ||
| 49 | NEXN | 12737 1903 | 11331 | -0.006 | 0.1861 | No | ||
| 50 | FEZ1 | 19520 3100 | 11341 | -0.006 | 0.1861 | No | ||
| 51 | PTEN | 5305 | 11437 | -0.007 | 0.1814 | No | ||
| 52 | SCG2 | 5410 14425 | 11790 | -0.008 | 0.1630 | No | ||
| 53 | OPHN1 | 24102 | 11990 | -0.009 | 0.1529 | No | ||
| 54 | CD24 | 8711 | 12061 | -0.009 | 0.1498 | No | ||
| 55 | EGFR | 1329 20944 | 12193 | -0.009 | 0.1434 | No | ||
| 56 | TDGF1 | 5704 10108 | 12335 | -0.010 | 0.1365 | No | ||
| 57 | TBX5 | 3531 5634 | 12427 | -0.010 | 0.1324 | No | ||
| 58 | SPON2 | 16565 | 12457 | -0.011 | 0.1316 | No | ||
| 59 | TNFSF12 | 10209 | 12485 | -0.011 | 0.1310 | No | ||
| 60 | SFTPD | 21867 | 12879 | -0.013 | 0.1107 | No | ||
| 61 | GLI2 | 13859 | 13099 | -0.014 | 0.0999 | No | ||
| 62 | BMP10 | 17376 | 13320 | -0.016 | 0.0893 | No | ||
| 63 | TRIP6 | 16331 | 13414 | -0.017 | 0.0855 | No | ||
| 64 | NRTN | 22923 | 14123 | -0.024 | 0.0490 | No | ||
| 65 | CDH13 | 4506 3826 | 14592 | -0.032 | 0.0261 | No | ||
| 66 | HMGCR | 9097 | 14745 | -0.035 | 0.0205 | No | ||
| 67 | SPHK1 | 5484 1266 1402 | 15081 | -0.043 | 0.0056 | No | ||
| 68 | CDK5R1 | 20745 | 15153 | -0.045 | 0.0051 | No | ||
| 69 | SCYE1 | 8897 | 15170 | -0.045 | 0.0076 | No | ||
| 70 | ENPEP | 4668 | 15351 | -0.051 | 0.0016 | No | ||
| 71 | IL8RB | 8752 4533 | 15434 | -0.054 | 0.0011 | No | ||
| 72 | FEZ2 | 22895 | 15714 | -0.066 | -0.0091 | No | ||
| 73 | ITGB1 | 3872 18411 | 15782 | -0.069 | -0.0076 | No | ||
| 74 | UBB | 10240 | 16120 | -0.086 | -0.0194 | No | ||
| 75 | DPYSL5 | 92 | 16432 | -0.109 | -0.0282 | No | ||
| 76 | CLIC4 | 11386 11342 6620 11387 | 17225 | -0.188 | -0.0571 | No | ||
| 77 | GTPBP4 | 21548 | 17228 | -0.188 | -0.0433 | No | ||
| 78 | VEGFC | 10286 | 17467 | -0.221 | -0.0399 | No | ||
| 79 | BCAR1 | 18741 | 18276 | -0.468 | -0.0490 | No | ||
| 80 | TGFB2 | 5743 | 18487 | -0.915 | 0.0070 | No |

