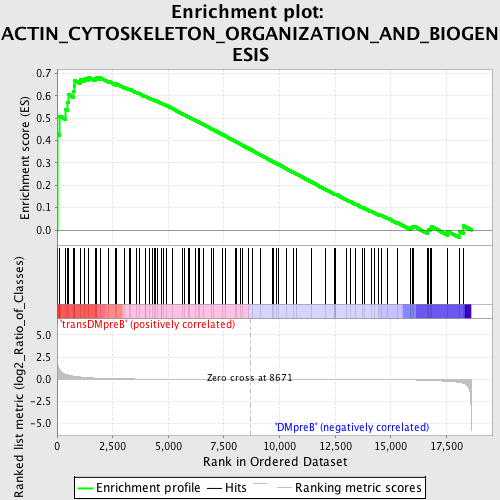
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | ACTIN_CYTOSKELETON_ORGANIZATION_AND_BIOGENESIS |
| Enrichment Score (ES) | 0.6821488 |
| Normalized Enrichment Score (NES) | 1.5632336 |
| Nominal p-value | 0.0019120459 |
| FDR q-value | 0.29089996 |
| FWER p-Value | 0.988 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MYH9 | 2252 2244 | 1 | 5.596 | 0.4299 | Yes | ||
| 2 | FGD2 | 23309 | 109 | 1.115 | 0.5098 | Yes | ||
| 3 | CAPG | 17411 | 371 | 0.565 | 0.5391 | Yes | ||
| 4 | ROCK1 | 5386 | 458 | 0.495 | 0.5725 | Yes | ||
| 5 | EVL | 21156 | 507 | 0.470 | 0.6061 | Yes | ||
| 6 | DOCK2 | 8243 | 718 | 0.354 | 0.6220 | Yes | ||
| 7 | WASL | 17203 | 770 | 0.333 | 0.6448 | Yes | ||
| 8 | FLNA | 24129 | 798 | 0.324 | 0.6682 | Yes | ||
| 9 | WASF2 | 6326 | 1054 | 0.248 | 0.6735 | Yes | ||
| 10 | ATP2C1 | 10660 6178 | 1251 | 0.202 | 0.6784 | Yes | ||
| 11 | RASA1 | 10174 | 1426 | 0.170 | 0.6821 | Yes | ||
| 12 | WIPF1 | 14560 | 1705 | 0.129 | 0.6770 | No | ||
| 13 | PDPK1 | 23097 | 1785 | 0.119 | 0.6819 | No | ||
| 14 | ARHGDIB | 16950 | 1928 | 0.100 | 0.6819 | No | ||
| 15 | FLNB | 13742 | 2324 | 0.066 | 0.6656 | No | ||
| 16 | ABL1 | 2693 4301 2794 | 2639 | 0.049 | 0.6524 | No | ||
| 17 | FSCN1 | 16637 | 2650 | 0.049 | 0.6556 | No | ||
| 18 | GHSR | 1810 15625 | 3029 | 0.036 | 0.6380 | No | ||
| 19 | TSC1 | 7235 | 3237 | 0.031 | 0.6292 | No | ||
| 20 | NF1 | 5165 | 3307 | 0.029 | 0.6277 | No | ||
| 21 | ARPC4 | 12642 | 3567 | 0.024 | 0.6155 | No | ||
| 22 | CXCL1 | 9044 | 3693 | 0.022 | 0.6105 | No | ||
| 23 | NF2 | 1222 5166 | 3971 | 0.018 | 0.5969 | No | ||
| 24 | RHOF | 16379 | 3989 | 0.018 | 0.5974 | No | ||
| 25 | SSH1 | 10540 | 4137 | 0.017 | 0.5907 | No | ||
| 26 | FGD3 | 11399 | 4302 | 0.015 | 0.5830 | No | ||
| 27 | RND3 | 13181 | 4367 | 0.014 | 0.5807 | No | ||
| 28 | FGD4 | 10306 5870 1717 1669 1700 | 4433 | 0.014 | 0.5782 | No | ||
| 29 | DST | 3934 3984 4644 3976 14281 4070 4020 4008 4040 | 4521 | 0.013 | 0.5746 | No | ||
| 30 | DLG1 | 22793 1672 | 4695 | 0.012 | 0.5662 | No | ||
| 31 | RAC3 | 20561 | 4699 | 0.012 | 0.5669 | No | ||
| 32 | LIMK1 | 16350 | 4780 | 0.012 | 0.5635 | No | ||
| 33 | PACSIN2 | 2312 | 4924 | 0.011 | 0.5566 | No | ||
| 34 | MRAS | 9409 | 5175 | 0.009 | 0.5438 | No | ||
| 35 | CDC42BPA | 10384 | 5654 | 0.007 | 0.5186 | No | ||
| 36 | PRKG1 | 5289 | 5731 | 0.007 | 0.5151 | No | ||
| 37 | CDC42 | 4503 8722 4504 2465 | 5905 | 0.007 | 0.5062 | No | ||
| 38 | TNXB | 1557 8174 878 8175 | 5971 | 0.006 | 0.5032 | No | ||
| 39 | NCK1 | 9447 5152 | 6199 | 0.006 | 0.4914 | No | ||
| 40 | CDC42EP5 | 17991 | 6350 | 0.005 | 0.4837 | No | ||
| 41 | CENTD2 | 1758 18169 2284 | 6381 | 0.005 | 0.4825 | No | ||
| 42 | RHOA | 8624 4409 4410 | 6567 | 0.005 | 0.4729 | No | ||
| 43 | WASF1 | 13514 | 6958 | 0.004 | 0.4521 | No | ||
| 44 | DSTN | 14823 | 7021 | 0.003 | 0.4490 | No | ||
| 45 | GSN | 2784 | 7437 | 0.003 | 0.4268 | No | ||
| 46 | CRK | 4559 1249 | 7552 | 0.002 | 0.4209 | No | ||
| 47 | DBN1 | 21452 | 8038 | 0.001 | 0.3948 | No | ||
| 48 | ACTA1 | 18715 | 8080 | 0.001 | 0.3927 | No | ||
| 49 | MTSS1 | 5585 22285 | 8257 | 0.001 | 0.3832 | No | ||
| 50 | SORBS3 | 21766 | 8314 | 0.001 | 0.3803 | No | ||
| 51 | FGD6 | 19901 | 8581 | 0.000 | 0.3659 | No | ||
| 52 | MYOZ1 | 21907 | 8777 | -0.000 | 0.3554 | No | ||
| 53 | PLEK2 | 21033 | 9133 | -0.001 | 0.3363 | No | ||
| 54 | FSCN2 | 20569 | 9674 | -0.002 | 0.3073 | No | ||
| 55 | FGD1 | 24228 | 9726 | -0.002 | 0.3048 | No | ||
| 56 | TESK2 | 10504 | 9878 | -0.003 | 0.2968 | No | ||
| 57 | TTN | 14552 2743 14553 | 9972 | -0.003 | 0.2920 | No | ||
| 58 | GHRL | 1183 17040 | 10313 | -0.004 | 0.2740 | No | ||
| 59 | MYH11 | 22656 | 10609 | -0.004 | 0.2584 | No | ||
| 60 | AMOT | 11341 24037 6587 | 10754 | -0.005 | 0.2510 | No | ||
| 61 | RACGAP1 | 2240 22132 | 11453 | -0.007 | 0.2138 | No | ||
| 62 | TAOK2 | 10610 3794 | 12059 | -0.009 | 0.1818 | No | ||
| 63 | ARHGEF17 | 17737 | 12483 | -0.011 | 0.1598 | No | ||
| 64 | ADRA2A | 4355 | 12489 | -0.011 | 0.1604 | No | ||
| 65 | NEBL | 14678 2691 | 12493 | -0.011 | 0.1611 | No | ||
| 66 | CDC42EP2 | 4180 14771 | 12523 | -0.011 | 0.1603 | No | ||
| 67 | PLA2G1B | 9581 | 13015 | -0.014 | 0.1349 | No | ||
| 68 | ARPC5 | 12580 7487 | 13183 | -0.015 | 0.1270 | No | ||
| 69 | ARHGEF2 | 4987 | 13419 | -0.017 | 0.1156 | No | ||
| 70 | RHOJ | 21241 | 13731 | -0.020 | 0.1004 | No | ||
| 71 | ELMO1 | 3275 8939 3202 3271 3254 3235 3164 3175 3274 3188 3293 3214 3218 3239 | 13808 | -0.021 | 0.0978 | No | ||
| 72 | WASF3 | 16625 | 14120 | -0.024 | 0.0829 | No | ||
| 73 | RND1 | 5868 | 14256 | -0.026 | 0.0776 | No | ||
| 74 | PSCD2 | 17826 | 14466 | -0.029 | 0.0686 | No | ||
| 75 | CXCL12 | 9792 1182 1031 | 14573 | -0.032 | 0.0653 | No | ||
| 76 | FGD5 | 10555 17352 | 14846 | -0.038 | 0.0535 | No | ||
| 77 | NCK2 | 9448 | 15281 | -0.049 | 0.0338 | No | ||
| 78 | SSH2 | 6202 | 15880 | -0.074 | 0.0072 | No | ||
| 79 | CFL1 | 4516 | 15903 | -0.075 | 0.0118 | No | ||
| 80 | SCIN | 21079 | 15974 | -0.078 | 0.0141 | No | ||
| 81 | ARFIP2 | 1226 17702 | 16025 | -0.081 | 0.0176 | No | ||
| 82 | ARF6 | 21252 | 16656 | -0.127 | -0.0066 | No | ||
| 83 | CDC42BPB | 20982 | 16673 | -0.129 | 0.0024 | No | ||
| 84 | KPTN | 18385 | 16762 | -0.137 | 0.0082 | No | ||
| 85 | RAC1 | 16302 | 16844 | -0.147 | 0.0151 | No | ||
| 86 | DYNLL1 | 12135 | 17550 | -0.231 | -0.0051 | No | ||
| 87 | ARPC1A | 3541 16628 3519 | 18073 | -0.346 | -0.0067 | No | ||
| 88 | BCAR1 | 18741 | 18276 | -0.468 | 0.0183 | No |

