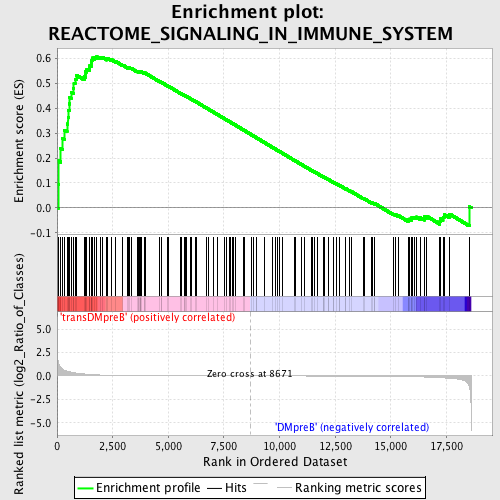
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | REACTOME_SIGNALING_IN_IMMUNE_SYSTEM |
| Enrichment Score (ES) | 0.6054788 |
| Normalized Enrichment Score (NES) | 1.4659194 |
| Nominal p-value | 0.0054151625 |
| FDR q-value | 0.9262703 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GRB7 | 20673 | 44 | 1.580 | 0.0956 | Yes | ||
| 2 | MAP3K7IP1 | 2193 2171 22419 | 59 | 1.517 | 0.1890 | Yes | ||
| 3 | CD86 | 1660 22601 | 165 | 0.896 | 0.2389 | Yes | ||
| 4 | FCER1G | 13759 | 239 | 0.726 | 0.2799 | Yes | ||
| 5 | CD47 | 4933 | 338 | 0.593 | 0.3114 | Yes | ||
| 6 | FCGR2B | 8959 | 449 | 0.503 | 0.3366 | Yes | ||
| 7 | PTPN6 | 17002 | 489 | 0.478 | 0.3642 | Yes | ||
| 8 | UBE2N | 8216 19898 | 515 | 0.466 | 0.3918 | Yes | ||
| 9 | WAS | 24193 | 554 | 0.440 | 0.4170 | Yes | ||
| 10 | C3 | 22915 | 569 | 0.432 | 0.4430 | Yes | ||
| 11 | MAP3K1 | 21348 | 633 | 0.393 | 0.4640 | Yes | ||
| 12 | CD19 | 17640 | 745 | 0.342 | 0.4792 | Yes | ||
| 13 | CD48 | 14051 | 758 | 0.337 | 0.4995 | Yes | ||
| 14 | TIRAP | 3075 19187 | 810 | 0.320 | 0.5166 | Yes | ||
| 15 | IKBKB | 4907 | 861 | 0.304 | 0.5327 | Yes | ||
| 16 | IRF3 | 2149 2188 18255 | 1221 | 0.207 | 0.5261 | Yes | ||
| 17 | CSK | 8805 | 1270 | 0.197 | 0.5357 | Yes | ||
| 18 | BSG | 19964 3331 | 1276 | 0.195 | 0.5476 | Yes | ||
| 19 | MAP3K7 | 16255 | 1335 | 0.182 | 0.5557 | Yes | ||
| 20 | NFKBIA | 21065 | 1447 | 0.167 | 0.5601 | Yes | ||
| 21 | SELL | 9798 | 1468 | 0.163 | 0.5691 | Yes | ||
| 22 | RELA | 23783 | 1532 | 0.154 | 0.5753 | Yes | ||
| 23 | MAP3K8 | 23495 | 1536 | 0.153 | 0.5846 | Yes | ||
| 24 | CHUK | 23665 | 1559 | 0.150 | 0.5928 | Yes | ||
| 25 | VAV1 | 23173 | 1577 | 0.148 | 0.6010 | Yes | ||
| 26 | TYROBP | 18302 | 1673 | 0.133 | 0.6041 | Yes | ||
| 27 | PDPK1 | 23097 | 1785 | 0.119 | 0.6055 | Yes | ||
| 28 | ITGAL | 9187 | 1927 | 0.100 | 0.6040 | No | ||
| 29 | ITGB2 | 19978 | 2033 | 0.089 | 0.6039 | No | ||
| 30 | CD3D | 19473 | 2239 | 0.072 | 0.5973 | No | ||
| 31 | PPIA | 1284 11188 | 2281 | 0.069 | 0.5993 | No | ||
| 32 | C1QB | 8667 | 2428 | 0.061 | 0.5952 | No | ||
| 33 | VCAM1 | 5851 | 2636 | 0.049 | 0.5870 | No | ||
| 34 | ZAP70 | 14271 4042 | 2953 | 0.038 | 0.5723 | No | ||
| 35 | C1S | 6950 6949 | 3178 | 0.032 | 0.5622 | No | ||
| 36 | PIK3C3 | 23607 | 3199 | 0.032 | 0.5631 | No | ||
| 37 | RIPK1 | 5381 | 3271 | 0.030 | 0.5611 | No | ||
| 38 | FYB | 10717 | 3333 | 0.028 | 0.5595 | No | ||
| 39 | MBL2 | 23886 | 3590 | 0.024 | 0.5471 | No | ||
| 40 | PAK1 | 9527 | 3642 | 0.023 | 0.5458 | No | ||
| 41 | LAT | 17643 | 3664 | 0.023 | 0.5460 | No | ||
| 42 | CFD | 19957 | 3702 | 0.022 | 0.5454 | No | ||
| 43 | CD160 | 12023 | 3727 | 0.021 | 0.5454 | No | ||
| 44 | ITGB3 | 20631 | 3749 | 0.021 | 0.5456 | No | ||
| 45 | RIPK2 | 2528 15935 | 3768 | 0.021 | 0.5459 | No | ||
| 46 | NFKBIB | 17906 | 3804 | 0.020 | 0.5453 | No | ||
| 47 | TLR1 | 10190 | 3934 | 0.019 | 0.5395 | No | ||
| 48 | CD14 | 23452 | 3948 | 0.019 | 0.5399 | No | ||
| 49 | DOK2 | 354 | 3951 | 0.019 | 0.5410 | No | ||
| 50 | LBP | 14760 | 4598 | 0.013 | 0.5068 | No | ||
| 51 | C6 | 22522 8671 | 4703 | 0.012 | 0.5019 | No | ||
| 52 | CAV1 | 8698 | 4710 | 0.012 | 0.5023 | No | ||
| 53 | ITGA5 | 22105 | 4968 | 0.010 | 0.4891 | No | ||
| 54 | CD28 | 14239 4092 | 4992 | 0.010 | 0.4885 | No | ||
| 55 | PLCG1 | 14753 | 5548 | 0.008 | 0.4589 | No | ||
| 56 | GRB14 | 14574 2719 | 5569 | 0.008 | 0.4583 | No | ||
| 57 | B2M | 72 14882 | 5721 | 0.007 | 0.4506 | No | ||
| 58 | SOS1 | 5476 | 5736 | 0.007 | 0.4503 | No | ||
| 59 | CD3G | 19139 | 5785 | 0.007 | 0.4481 | No | ||
| 60 | ITGAX | 18058 | 5823 | 0.007 | 0.4466 | No | ||
| 61 | PTPRC | 5327 9662 | 5983 | 0.006 | 0.4384 | No | ||
| 62 | ITGAV | 4931 | 6031 | 0.006 | 0.4362 | No | ||
| 63 | NCK1 | 9447 5152 | 6199 | 0.006 | 0.4275 | No | ||
| 64 | LCP2 | 4988 9268 | 6272 | 0.005 | 0.4240 | No | ||
| 65 | ANGPT1 | 8588 2260 22303 | 6701 | 0.004 | 0.4011 | No | ||
| 66 | CXADR | 22720 | 6818 | 0.004 | 0.3950 | No | ||
| 67 | C8A | 15823 | 7044 | 0.003 | 0.3831 | No | ||
| 68 | ICOSLG | 19973 | 7202 | 0.003 | 0.3748 | No | ||
| 69 | GAS6 | 18927 | 7527 | 0.002 | 0.3574 | No | ||
| 70 | SPN | 452 5493 | 7595 | 0.002 | 0.3539 | No | ||
| 71 | C8B | 16163 | 7731 | 0.002 | 0.3467 | No | ||
| 72 | PIK3R1 | 3170 | 7781 | 0.002 | 0.3442 | No | ||
| 73 | OLR1 | 8427 4233 | 7897 | 0.002 | 0.3380 | No | ||
| 74 | EEA1 | 5695 10097 | 7921 | 0.001 | 0.3369 | No | ||
| 75 | ITK | 4934 | 8022 | 0.001 | 0.3316 | No | ||
| 76 | LYN | 16281 | 8372 | 0.001 | 0.3127 | No | ||
| 77 | CDH1 | 18479 | 8402 | 0.001 | 0.3112 | No | ||
| 78 | KIR3DL1 | 2611 6365 | 8747 | -0.000 | 0.2926 | No | ||
| 79 | CD40LG | 24330 | 8842 | -0.000 | 0.2875 | No | ||
| 80 | ITGA4 | 4929 | 8961 | -0.001 | 0.2812 | No | ||
| 81 | TLR5 | 14017 | 8983 | -0.001 | 0.2801 | No | ||
| 82 | PRKCQ | 2873 2831 | 9311 | -0.001 | 0.2624 | No | ||
| 83 | AKT1 | 8568 | 9661 | -0.002 | 0.2437 | No | ||
| 84 | C9 | 22518 2246 | 9802 | -0.002 | 0.2363 | No | ||
| 85 | ENAH | 4665 8901 | 9896 | -0.003 | 0.2314 | No | ||
| 86 | CD3E | 8714 | 10010 | -0.003 | 0.2255 | No | ||
| 87 | MASP1 | 1632 22626 | 10112 | -0.003 | 0.2202 | No | ||
| 88 | ANGPT2 | 14885 3783 18923 | 10658 | -0.005 | 0.1910 | No | ||
| 89 | TICAM2 | 23440 | 10724 | -0.005 | 0.1878 | No | ||
| 90 | GRAP2 | 5113 9398 | 10970 | -0.005 | 0.1749 | No | ||
| 91 | CRTAM | 19160 | 11108 | -0.006 | 0.1678 | No | ||
| 92 | PTEN | 5305 | 11437 | -0.007 | 0.1505 | No | ||
| 93 | PIK3CA | 9562 | 11488 | -0.007 | 0.1482 | No | ||
| 94 | PROC | 23475 | 11587 | -0.007 | 0.1433 | No | ||
| 95 | TEK | 16175 | 11713 | -0.008 | 0.1370 | No | ||
| 96 | IFITM1 | 18017 7578 12724 | 11959 | -0.008 | 0.1243 | No | ||
| 97 | JAM2 | 7428 22718 12519 | 11964 | -0.008 | 0.1246 | No | ||
| 98 | PTPN11 | 5326 16391 9660 | 12011 | -0.009 | 0.1226 | No | ||
| 99 | CD226 | 5920 | 12185 | -0.009 | 0.1139 | No | ||
| 100 | CD4 | 16999 | 12192 | -0.009 | 0.1141 | No | ||
| 101 | CD96 | 22580 | 12400 | -0.010 | 0.1036 | No | ||
| 102 | CTLA4 | 14238 | 12444 | -0.011 | 0.1019 | No | ||
| 103 | ITGAM | 9188 | 12542 | -0.011 | 0.0973 | No | ||
| 104 | C1QA | 8666 | 12698 | -0.012 | 0.0897 | No | ||
| 105 | C1R | 11942 6951 6952 | 12702 | -0.012 | 0.0902 | No | ||
| 106 | MERTK | 5094 | 12975 | -0.014 | 0.0764 | No | ||
| 107 | TLR3 | 18884 | 12979 | -0.014 | 0.0771 | No | ||
| 108 | SHC1 | 9813 9812 5430 | 13160 | -0.015 | 0.0682 | No | ||
| 109 | C1QC | 15706 | 13243 | -0.015 | 0.0648 | No | ||
| 110 | APOB | 21322 | 13759 | -0.020 | 0.0381 | No | ||
| 111 | NFKB1 | 15160 | 13838 | -0.021 | 0.0352 | No | ||
| 112 | TLR4 | 2329 10191 5770 | 14147 | -0.024 | 0.0200 | No | ||
| 113 | PDCD1 | 13870 | 14171 | -0.025 | 0.0203 | No | ||
| 114 | MAG | 17880 | 14274 | -0.026 | 0.0164 | No | ||
| 115 | SLC16A3 | 481 482 | 14284 | -0.026 | 0.0176 | No | ||
| 116 | FN1 | 4091 4094 971 13929 | 15113 | -0.044 | -0.0245 | No | ||
| 117 | ATF2 | 4418 2759 | 15227 | -0.047 | -0.0277 | No | ||
| 118 | CD200 | 9404 58 | 15348 | -0.051 | -0.0310 | No | ||
| 119 | ITGB1 | 3872 18411 | 15782 | -0.069 | -0.0502 | No | ||
| 120 | FYN | 3375 3395 20052 | 15795 | -0.069 | -0.0465 | No | ||
| 121 | CD2 | 15223 | 15836 | -0.071 | -0.0443 | No | ||
| 122 | CD80 | 22758 | 15922 | -0.076 | -0.0442 | No | ||
| 123 | KRAS | 9247 | 15933 | -0.076 | -0.0400 | No | ||
| 124 | LY86 | 21665 | 15965 | -0.078 | -0.0368 | No | ||
| 125 | F2 | 14524 | 16051 | -0.082 | -0.0363 | No | ||
| 126 | C2 | 23013 | 16152 | -0.089 | -0.0362 | No | ||
| 127 | PPIL2 | 22648 | 16319 | -0.100 | -0.0390 | No | ||
| 128 | MAPK8 | 6459 | 16496 | -0.113 | -0.0415 | No | ||
| 129 | JUN | 15832 | 16520 | -0.114 | -0.0357 | No | ||
| 130 | TLR2 | 15308 | 16615 | -0.123 | -0.0331 | No | ||
| 131 | PIK3CB | 19030 | 17205 | -0.186 | -0.0535 | No | ||
| 132 | THBD | 14404 | 17239 | -0.190 | -0.0435 | No | ||
| 133 | BCL10 | 15397 | 17346 | -0.203 | -0.0366 | No | ||
| 134 | CD81 | 8719 | 17396 | -0.209 | -0.0262 | No | ||
| 135 | MAPK1 | 1642 11167 | 17642 | -0.244 | -0.0243 | No | ||
| 136 | LCK | 15746 | 18528 | -1.241 | 0.0048 | No |

