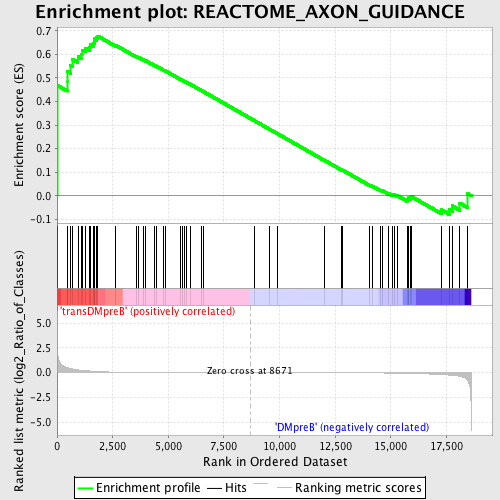
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | REACTOME_AXON_GUIDANCE |
| Enrichment Score (ES) | 0.6775431 |
| Normalized Enrichment Score (NES) | 1.4418156 |
| Nominal p-value | 0.031135531 |
| FDR q-value | 0.890348 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MYH9 | 2252 2244 | 1 | 5.596 | 0.4690 | Yes | ||
| 2 | AGRN | 4361 | 455 | 0.496 | 0.4861 | Yes | ||
| 3 | ROCK1 | 5386 | 458 | 0.495 | 0.5275 | Yes | ||
| 4 | HSP90AB1 | 4880 9130 4881 | 606 | 0.408 | 0.5538 | Yes | ||
| 5 | ARHGEF11 | 10028 | 701 | 0.361 | 0.5790 | Yes | ||
| 6 | RHOG | 17727 | 938 | 0.280 | 0.5897 | Yes | ||
| 7 | RRAS | 18256 | 1108 | 0.230 | 0.5999 | Yes | ||
| 8 | MAP2K1 | 19082 | 1120 | 0.227 | 0.6184 | Yes | ||
| 9 | MAPK3 | 6458 11170 | 1268 | 0.198 | 0.6270 | Yes | ||
| 10 | RAF1 | 17035 | 1454 | 0.166 | 0.6309 | Yes | ||
| 11 | YWHAB | 14744 | 1492 | 0.160 | 0.6423 | Yes | ||
| 12 | CDK5 | 16591 | 1632 | 0.139 | 0.6465 | Yes | ||
| 13 | TYROBP | 18302 | 1673 | 0.133 | 0.6555 | Yes | ||
| 14 | PTK2 | 22271 | 1691 | 0.130 | 0.6655 | Yes | ||
| 15 | PTPRA | 14844 | 1784 | 0.119 | 0.6705 | Yes | ||
| 16 | MAP2K2 | 19933 | 1831 | 0.114 | 0.6775 | Yes | ||
| 17 | ABL1 | 2693 4301 2794 | 2639 | 0.049 | 0.6382 | No | ||
| 18 | CLASP2 | 13338 | 3558 | 0.024 | 0.5907 | No | ||
| 19 | PAK1 | 9527 | 3642 | 0.023 | 0.5882 | No | ||
| 20 | PIP5K1C | 5250 19929 | 3871 | 0.020 | 0.5775 | No | ||
| 21 | CREB1 | 3990 8782 4558 4093 | 3985 | 0.018 | 0.5729 | No | ||
| 22 | NCAM1 | 5149 | 4368 | 0.014 | 0.5536 | No | ||
| 23 | SEMA3A | 16919 | 4448 | 0.014 | 0.5505 | No | ||
| 24 | LIMK1 | 16350 | 4780 | 0.012 | 0.5336 | No | ||
| 25 | RPS6KA5 | 2076 21005 | 4851 | 0.011 | 0.5308 | No | ||
| 26 | DPYSL2 | 3122 4561 8787 | 5559 | 0.008 | 0.4933 | No | ||
| 27 | PLXNC1 | 7056 | 5618 | 0.008 | 0.4908 | No | ||
| 28 | SOS1 | 5476 | 5736 | 0.007 | 0.4852 | No | ||
| 29 | SEMA7A | 19426 9803 | 5814 | 0.007 | 0.4816 | No | ||
| 30 | PTPRC | 5327 9662 | 5983 | 0.006 | 0.4731 | No | ||
| 31 | SLIT2 | 5456 | 6511 | 0.005 | 0.4451 | No | ||
| 32 | RHOA | 8624 4409 4410 | 6567 | 0.005 | 0.4425 | No | ||
| 33 | CNTN2 | 5627 | 8878 | -0.000 | 0.3181 | No | ||
| 34 | CAP2 | 21649 | 9554 | -0.002 | 0.2819 | No | ||
| 35 | SRGAP2 | 13836 17045 6433 17046 | 9901 | -0.003 | 0.2635 | No | ||
| 36 | SEMA3E | 16918 | 12002 | -0.009 | 0.1510 | No | ||
| 37 | NCAN | 3898 10164 8810 4570 18856 | 12787 | -0.012 | 0.1098 | No | ||
| 38 | FES | 17779 3949 | 12817 | -0.013 | 0.1093 | No | ||
| 39 | MET | 17520 | 14045 | -0.023 | 0.0452 | No | ||
| 40 | ERBB2 | 8913 | 14185 | -0.025 | 0.0397 | No | ||
| 41 | FARP2 | 14175 | 14530 | -0.031 | 0.0238 | No | ||
| 42 | PSPN | 22921 | 14609 | -0.032 | 0.0223 | No | ||
| 43 | SEMA5A | 22496 5423 | 14904 | -0.039 | 0.0097 | No | ||
| 44 | SEMA6D | 2800 14876 90 | 15085 | -0.043 | 0.0036 | No | ||
| 45 | CDK5R1 | 20745 | 15153 | -0.045 | 0.0038 | No | ||
| 46 | NCK2 | 9448 | 15281 | -0.049 | 0.0011 | No | ||
| 47 | CD72 | 8718 | 15729 | -0.066 | -0.0174 | No | ||
| 48 | ITGB1 | 3872 18411 | 15782 | -0.069 | -0.0145 | No | ||
| 49 | FYN | 3375 3395 20052 | 15795 | -0.069 | -0.0093 | No | ||
| 50 | CFL1 | 4516 | 15903 | -0.075 | -0.0088 | No | ||
| 51 | KRAS | 9247 | 15933 | -0.076 | -0.0039 | No | ||
| 52 | PRNP | 9622 | 17277 | -0.195 | -0.0600 | No | ||
| 53 | MAPK1 | 1642 11167 | 17642 | -0.244 | -0.0591 | No | ||
| 54 | MYL6 | 9438 3408 | 17758 | -0.266 | -0.0430 | No | ||
| 55 | GPC1 | 4796 | 18103 | -0.357 | -0.0316 | No | ||
| 56 | GFRA1 | 9015 | 18437 | -0.707 | 0.0096 | No |

