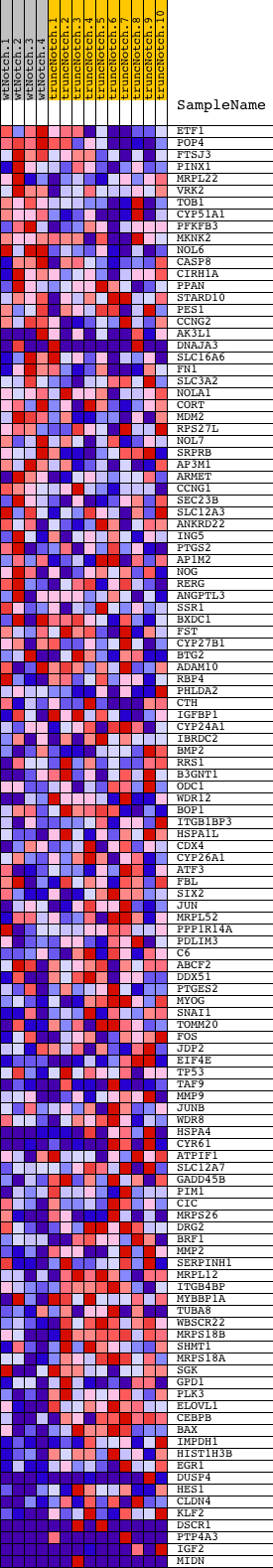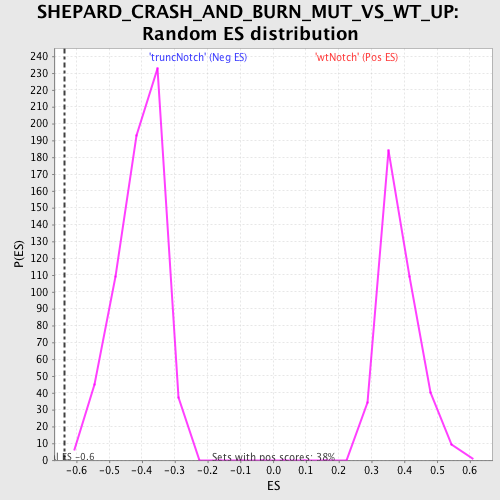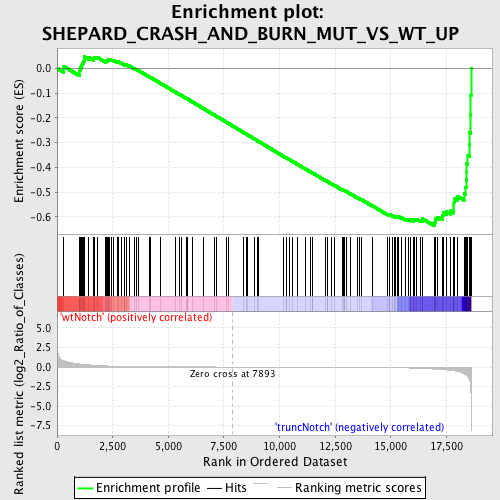
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_truncNotch.phenotype_wtNotch_versus_truncNotch.cls #wtNotch_versus_truncNotch.phenotype_wtNotch_versus_truncNotch.cls #wtNotch_versus_truncNotch_repos |
| Phenotype | phenotype_wtNotch_versus_truncNotch.cls#wtNotch_versus_truncNotch_repos |
| Upregulated in class | truncNotch |
| GeneSet | SHEPARD_CRASH_AND_BURN_MUT_VS_WT_UP |
| Enrichment Score (ES) | -0.6369492 |
| Normalized Enrichment Score (NES) | -1.5699059 |
| Nominal p-value | 0.0016051364 |
| FDR q-value | 0.25785804 |
| FWER p-Value | 0.996 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ETF1 | 6770075 | 307 | 0.780 | 0.0076 | No | ||
| 2 | POP4 | 4560600 | 1015 | 0.388 | -0.0185 | No | ||
| 3 | FTSJ3 | 3140100 | 1016 | 0.388 | -0.0065 | No | ||
| 4 | PINX1 | 3360154 | 1055 | 0.372 | 0.0030 | No | ||
| 5 | MRPL22 | 6400520 | 1105 | 0.357 | 0.0115 | No | ||
| 6 | VRK2 | 3800692 6940056 | 1119 | 0.354 | 0.0218 | No | ||
| 7 | TOB1 | 4150138 | 1181 | 0.340 | 0.0291 | No | ||
| 8 | CYP51A1 | 4590427 | 1233 | 0.327 | 0.0365 | No | ||
| 9 | PFKFB3 | 630706 | 1234 | 0.327 | 0.0466 | No | ||
| 10 | MKNK2 | 1240020 5270092 | 1413 | 0.275 | 0.0456 | No | ||
| 11 | NOL6 | 2810504 3120050 3840021 6860315 | 1657 | 0.224 | 0.0394 | No | ||
| 12 | CASP8 | 4850154 | 1665 | 0.223 | 0.0460 | No | ||
| 13 | CIRH1A | 2340372 | 1813 | 0.196 | 0.0441 | No | ||
| 14 | PPAN | 540398 | 2179 | 0.141 | 0.0288 | No | ||
| 15 | STARD10 | 1400619 4570170 | 2221 | 0.135 | 0.0308 | No | ||
| 16 | PES1 | 2470427 | 2279 | 0.129 | 0.0317 | No | ||
| 17 | CCNG2 | 3190095 | 2287 | 0.128 | 0.0353 | No | ||
| 18 | AK3L1 | 7100601 | 2356 | 0.120 | 0.0354 | No | ||
| 19 | DNAJA3 | 1980671 6220338 | 2454 | 0.111 | 0.0335 | No | ||
| 20 | SLC16A6 | 1690156 | 2524 | 0.104 | 0.0330 | No | ||
| 21 | FN1 | 1170601 2970647 6220288 6940037 | 2696 | 0.090 | 0.0266 | No | ||
| 22 | SLC3A2 | 5270358 | 2740 | 0.086 | 0.0269 | No | ||
| 23 | NOLA1 | 730093 | 2910 | 0.074 | 0.0201 | No | ||
| 24 | CORT | 360008 2340092 | 3036 | 0.066 | 0.0154 | No | ||
| 25 | MDM2 | 3450053 5080138 | 3045 | 0.065 | 0.0170 | No | ||
| 26 | RPS27L | 50577 | 3102 | 0.061 | 0.0158 | No | ||
| 27 | NOL7 | 6520332 | 3231 | 0.053 | 0.0106 | No | ||
| 28 | SRPRB | 1500138 5130156 5910064 | 3474 | 0.043 | -0.0012 | No | ||
| 29 | AP3M1 | 5340433 | 3573 | 0.039 | -0.0053 | No | ||
| 30 | ARMET | 5130670 | 3648 | 0.036 | -0.0081 | No | ||
| 31 | CCNG1 | 870402 1500685 6110594 | 4162 | 0.023 | -0.0352 | No | ||
| 32 | SEC23B | 3390632 3840497 | 4192 | 0.023 | -0.0360 | No | ||
| 33 | SLC12A3 | 1980592 2350056 | 4629 | 0.016 | -0.0591 | No | ||
| 34 | ANKRD22 | 4480746 | 5315 | 0.010 | -0.0958 | No | ||
| 35 | ING5 | 3830440 5910239 6200471 6650576 | 5497 | 0.009 | -0.1053 | No | ||
| 36 | PTGS2 | 2510301 3170369 | 5586 | 0.009 | -0.1098 | No | ||
| 37 | AP1M2 | 2900138 6040324 6370537 | 5819 | 0.007 | -0.1221 | No | ||
| 38 | NOG | 2320609 | 5847 | 0.007 | -0.1234 | No | ||
| 39 | RERG | 5910324 | 5880 | 0.007 | -0.1249 | No | ||
| 40 | ANGPTL3 | 6180497 | 6083 | 0.006 | -0.1356 | No | ||
| 41 | SSR1 | 5890100 | 6593 | 0.004 | -0.1630 | No | ||
| 42 | BXDC1 | 2640068 | 7064 | 0.002 | -0.1883 | No | ||
| 43 | FST | 1110600 | 7185 | 0.002 | -0.1948 | No | ||
| 44 | CYP27B1 | 5270114 5360010 | 7596 | 0.001 | -0.2169 | No | ||
| 45 | BTG2 | 2350411 | 7706 | 0.001 | -0.2228 | No | ||
| 46 | ADAM10 | 3780156 | 8393 | -0.001 | -0.2598 | No | ||
| 47 | RBP4 | 4540131 | 8528 | -0.002 | -0.2670 | No | ||
| 48 | PHLDA2 | 4810494 | 8570 | -0.002 | -0.2692 | No | ||
| 49 | CTH | 1190332 5290139 | 8861 | -0.003 | -0.2848 | No | ||
| 50 | IGFBP1 | 5720435 | 9025 | -0.003 | -0.2935 | No | ||
| 51 | CYP24A1 | 2340338 | 9065 | -0.003 | -0.2955 | No | ||
| 52 | IBRDC2 | 5130041 | 10160 | -0.007 | -0.3544 | No | ||
| 53 | BMP2 | 6620687 | 10310 | -0.007 | -0.3623 | No | ||
| 54 | RRS1 | 4560551 | 10321 | -0.007 | -0.3626 | No | ||
| 55 | B3GNT1 | 2350446 | 10453 | -0.008 | -0.3694 | No | ||
| 56 | ODC1 | 5670168 | 10563 | -0.008 | -0.3750 | No | ||
| 57 | WDR12 | 4560722 | 10818 | -0.009 | -0.3885 | No | ||
| 58 | BOP1 | 4810040 | 11149 | -0.011 | -0.4060 | No | ||
| 59 | ITGB1BP3 | 60441 730750 | 11394 | -0.012 | -0.4188 | No | ||
| 60 | HSPA1L | 4010538 | 11468 | -0.012 | -0.4224 | No | ||
| 61 | CDX4 | 6380164 | 11493 | -0.012 | -0.4233 | No | ||
| 62 | CYP26A1 | 380022 2690600 | 12057 | -0.016 | -0.4532 | No | ||
| 63 | ATF3 | 1940546 | 12131 | -0.017 | -0.4567 | No | ||
| 64 | FBL | 5130020 | 12165 | -0.017 | -0.4579 | No | ||
| 65 | SIX2 | 1450551 | 12315 | -0.018 | -0.4654 | No | ||
| 66 | JUN | 840170 | 12454 | -0.019 | -0.4723 | No | ||
| 67 | MRPL52 | 1170204 | 12847 | -0.023 | -0.4927 | No | ||
| 68 | PPP1R14A | 870114 4760187 | 12875 | -0.024 | -0.4935 | No | ||
| 69 | PDLIM3 | 1410095 | 12906 | -0.024 | -0.4943 | No | ||
| 70 | C6 | 1770154 2900129 | 12927 | -0.024 | -0.4947 | No | ||
| 71 | ABCF2 | 630040 3140035 6450601 | 13010 | -0.025 | -0.4983 | No | ||
| 72 | DDX51 | 5550044 6350270 6590750 | 13196 | -0.027 | -0.5075 | No | ||
| 73 | PTGES2 | 4210097 | 13497 | -0.032 | -0.5227 | No | ||
| 74 | MYOG | 3190672 | 13605 | -0.034 | -0.5275 | No | ||
| 75 | SNAI1 | 6590253 | 13687 | -0.035 | -0.5307 | No | ||
| 76 | TOMM20 | 6510113 | 14187 | -0.045 | -0.5563 | No | ||
| 77 | FOS | 1850315 | 14838 | -0.065 | -0.5894 | No | ||
| 78 | JDP2 | 2360500 | 14937 | -0.069 | -0.5926 | No | ||
| 79 | EIF4E | 1580403 70133 6380215 | 14939 | -0.069 | -0.5905 | No | ||
| 80 | TP53 | 6130707 | 15076 | -0.075 | -0.5955 | No | ||
| 81 | TAF9 | 1850458 3830136 | 15180 | -0.080 | -0.5986 | No | ||
| 82 | MMP9 | 580338 | 15215 | -0.082 | -0.5979 | No | ||
| 83 | JUNB | 4230048 | 15318 | -0.088 | -0.6007 | No | ||
| 84 | WDR8 | 520377 6200093 | 15342 | -0.089 | -0.5991 | No | ||
| 85 | HSPA4 | 1050170 3450309 | 15472 | -0.098 | -0.6030 | No | ||
| 86 | CYR61 | 1240408 5290026 4120452 6550008 | 15657 | -0.109 | -0.6096 | No | ||
| 87 | ATPIF1 | 7050577 | 15777 | -0.119 | -0.6123 | No | ||
| 88 | SLC12A7 | 6860079 | 15862 | -0.127 | -0.6129 | No | ||
| 89 | GADD45B | 2350408 | 15876 | -0.128 | -0.6096 | No | ||
| 90 | PIM1 | 630047 | 16020 | -0.143 | -0.6129 | No | ||
| 91 | CIC | 6370161 | 16078 | -0.149 | -0.6114 | No | ||
| 92 | MRPS26 | 4120113 | 16131 | -0.154 | -0.6094 | No | ||
| 93 | DRG2 | 6770563 | 16316 | -0.177 | -0.6138 | No | ||
| 94 | BRF1 | 2100047 | 16406 | -0.189 | -0.6128 | No | ||
| 95 | MMP2 | 2570603 | 16423 | -0.190 | -0.6077 | No | ||
| 96 | SERPINH1 | 6130014 | 16965 | -0.267 | -0.6286 | Yes | ||
| 97 | MRPL12 | 3120112 | 16980 | -0.270 | -0.6210 | Yes | ||
| 98 | ITGB4BP | 1400541 | 16994 | -0.272 | -0.6132 | Yes | ||
| 99 | MYBBP1A | 110592 2570435 2900711 | 17022 | -0.277 | -0.6061 | Yes | ||
| 100 | TUBA8 | 3710484 | 17103 | -0.291 | -0.6014 | Yes | ||
| 101 | WBSCR22 | 3130070 4120551 5570433 | 17310 | -0.333 | -0.6021 | Yes | ||
| 102 | MRPS18B | 5570086 | 17340 | -0.340 | -0.5931 | Yes | ||
| 103 | SHMT1 | 5900132 | 17352 | -0.342 | -0.5831 | Yes | ||
| 104 | MRPS18A | 6770433 | 17486 | -0.366 | -0.5789 | Yes | ||
| 105 | SGK | 1400131 2480056 | 17670 | -0.418 | -0.5758 | Yes | ||
| 106 | GPD1 | 2480095 | 17833 | -0.474 | -0.5698 | Yes | ||
| 107 | PLK3 | 2640592 | 17834 | -0.475 | -0.5550 | Yes | ||
| 108 | ELOVL1 | 2450019 4760138 5340685 | 17838 | -0.478 | -0.5403 | Yes | ||
| 109 | CEBPB | 2970019 | 17856 | -0.485 | -0.5261 | Yes | ||
| 110 | BAX | 3830008 | 18003 | -0.553 | -0.5168 | Yes | ||
| 111 | IMPDH1 | 3190735 7050546 | 18294 | -0.853 | -0.5060 | Yes | ||
| 112 | HIST1H3B | 4780092 | 18364 | -0.956 | -0.4800 | Yes | ||
| 113 | EGR1 | 4610347 | 18402 | -1.010 | -0.4506 | Yes | ||
| 114 | DUSP4 | 2690044 2850593 | 18417 | -1.050 | -0.4187 | Yes | ||
| 115 | HES1 | 4810280 | 18420 | -1.054 | -0.3861 | Yes | ||
| 116 | CLDN4 | 4920739 | 18465 | -1.162 | -0.3523 | Yes | ||
| 117 | KLF2 | 6860270 | 18525 | -1.525 | -0.3081 | Yes | ||
| 118 | DSCR1 | 4540048 6220039 | 18535 | -1.573 | -0.2596 | Yes | ||
| 119 | PTP4A3 | 3780504 6400364 | 18580 | -2.374 | -0.1882 | Yes | ||
| 120 | IGF2 | 6510020 | 18590 | -2.564 | -0.1090 | Yes | ||
| 121 | MIDN | 2260132 5220377 | 18606 | -3.548 | 0.0005 | Yes |