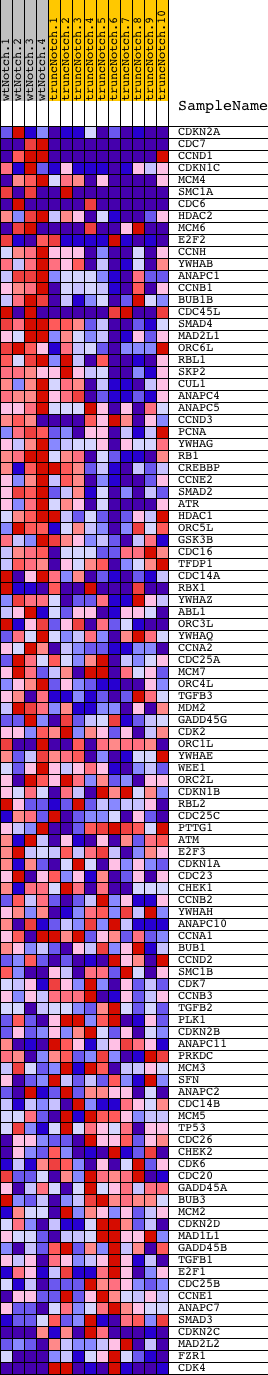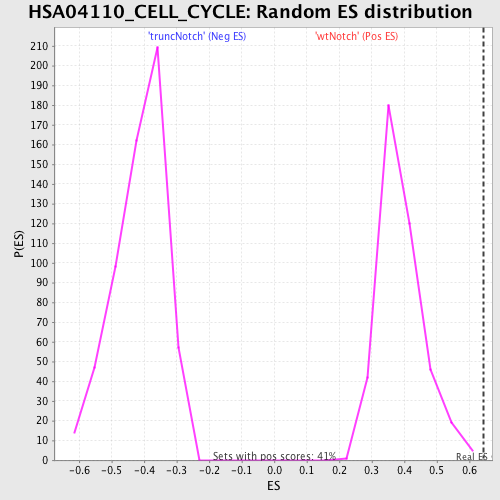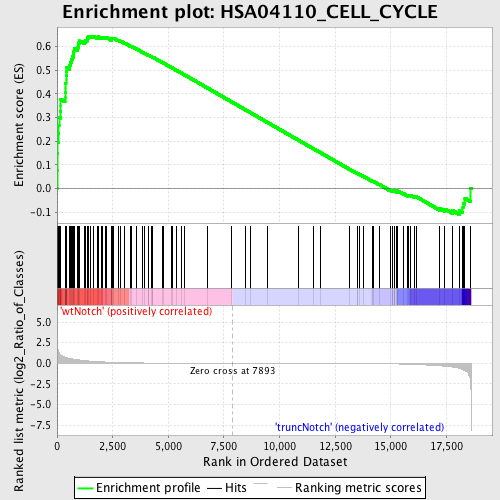
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_truncNotch.phenotype_wtNotch_versus_truncNotch.cls #wtNotch_versus_truncNotch.phenotype_wtNotch_versus_truncNotch.cls #wtNotch_versus_truncNotch_repos |
| Phenotype | phenotype_wtNotch_versus_truncNotch.cls#wtNotch_versus_truncNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | HSA04110_CELL_CYCLE |
| Enrichment Score (ES) | 0.64367795 |
| Normalized Enrichment Score (NES) | 1.6575499 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.14872952 |
| FWER p-Value | 0.746 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CDKN2A | 4670215 4760047 | 4 | 2.884 | 0.0765 | Yes | ||
| 2 | CDC7 | 4060546 4850041 | 7 | 2.689 | 0.1479 | Yes | ||
| 3 | CCND1 | 460524 770309 3120576 6980398 | 27 | 1.801 | 0.1947 | Yes | ||
| 4 | CDKN1C | 6520577 | 59 | 1.527 | 0.2337 | Yes | ||
| 5 | MCM4 | 2760673 5420711 | 78 | 1.324 | 0.2679 | Yes | ||
| 6 | SMC1A | 3060600 5700148 5890113 6370154 | 87 | 1.261 | 0.3010 | Yes | ||
| 7 | CDC6 | 4570296 5360600 | 145 | 1.030 | 0.3253 | Yes | ||
| 8 | HDAC2 | 4050433 | 161 | 0.999 | 0.3510 | Yes | ||
| 9 | MCM6 | 60092 540181 6510110 | 163 | 0.990 | 0.3773 | Yes | ||
| 10 | E2F2 | 5270609 5570377 7000465 | 356 | 0.731 | 0.3863 | Yes | ||
| 11 | CCNH | 3190347 | 360 | 0.727 | 0.4055 | Yes | ||
| 12 | YWHAB | 1740176 | 365 | 0.719 | 0.4244 | Yes | ||
| 13 | ANAPC1 | 6350162 | 366 | 0.719 | 0.4435 | Yes | ||
| 14 | CCNB1 | 4590433 4780372 | 400 | 0.685 | 0.4600 | Yes | ||
| 15 | BUB1B | 1450288 | 401 | 0.685 | 0.4782 | Yes | ||
| 16 | CDC45L | 70537 3130114 | 423 | 0.667 | 0.4948 | Yes | ||
| 17 | SMAD4 | 5670519 | 437 | 0.658 | 0.5116 | Yes | ||
| 18 | MAD2L1 | 4480725 | 569 | 0.583 | 0.5200 | Yes | ||
| 19 | ORC6L | 2260592 | 600 | 0.564 | 0.5334 | Yes | ||
| 20 | RBL1 | 3130372 | 638 | 0.543 | 0.5458 | Yes | ||
| 21 | SKP2 | 360711 380093 4810368 | 685 | 0.521 | 0.5572 | Yes | ||
| 22 | CUL1 | 1990632 | 723 | 0.503 | 0.5686 | Yes | ||
| 23 | ANAPC4 | 4540338 | 755 | 0.487 | 0.5798 | Yes | ||
| 24 | ANAPC5 | 730164 7100685 | 762 | 0.485 | 0.5924 | Yes | ||
| 25 | CCND3 | 380528 730500 | 912 | 0.426 | 0.5957 | Yes | ||
| 26 | PCNA | 940754 | 954 | 0.414 | 0.6045 | Yes | ||
| 27 | YWHAG | 3780341 | 966 | 0.409 | 0.6148 | Yes | ||
| 28 | RB1 | 5900338 | 1003 | 0.394 | 0.6233 | Yes | ||
| 29 | CREBBP | 5690035 7040050 | 1235 | 0.327 | 0.6195 | Yes | ||
| 30 | CCNE2 | 3120537 | 1281 | 0.315 | 0.6254 | Yes | ||
| 31 | SMAD2 | 4200592 | 1345 | 0.295 | 0.6299 | Yes | ||
| 32 | ATR | 6860273 | 1357 | 0.292 | 0.6370 | Yes | ||
| 33 | HDAC1 | 2850670 | 1396 | 0.280 | 0.6424 | Yes | ||
| 34 | ORC5L | 1940133 1940711 | 1519 | 0.252 | 0.6425 | Yes | ||
| 35 | GSK3B | 5360348 | 1613 | 0.232 | 0.6437 | Yes | ||
| 36 | CDC16 | 1940706 | 1827 | 0.194 | 0.6373 | No | ||
| 37 | TFDP1 | 1980112 | 1860 | 0.187 | 0.6406 | No | ||
| 38 | CDC14A | 4050132 | 2011 | 0.167 | 0.6369 | No | ||
| 39 | RBX1 | 2120010 2340047 | 2049 | 0.160 | 0.6392 | No | ||
| 40 | YWHAZ | 1230717 | 2155 | 0.145 | 0.6374 | No | ||
| 41 | ABL1 | 1050593 2030050 4010114 | 2206 | 0.138 | 0.6383 | No | ||
| 42 | ORC3L | 3060348 | 2434 | 0.113 | 0.6290 | No | ||
| 43 | YWHAQ | 6760524 | 2442 | 0.112 | 0.6316 | No | ||
| 44 | CCNA2 | 5290075 | 2458 | 0.110 | 0.6338 | No | ||
| 45 | CDC25A | 3800184 | 2485 | 0.107 | 0.6352 | No | ||
| 46 | MCM7 | 3290292 5220056 | 2530 | 0.103 | 0.6356 | No | ||
| 47 | ORC4L | 4230538 5550288 | 2753 | 0.085 | 0.6258 | No | ||
| 48 | TGFB3 | 1070041 | 2869 | 0.077 | 0.6217 | No | ||
| 49 | MDM2 | 3450053 5080138 | 3045 | 0.065 | 0.6139 | No | ||
| 50 | GADD45G | 2510142 | 3315 | 0.049 | 0.6007 | No | ||
| 51 | CDK2 | 130484 2260301 4010088 5050110 | 3331 | 0.049 | 0.6012 | No | ||
| 52 | ORC1L | 2370328 6110390 | 3561 | 0.039 | 0.5899 | No | ||
| 53 | YWHAE | 5310435 | 3837 | 0.030 | 0.5759 | No | ||
| 54 | WEE1 | 3390070 | 3945 | 0.027 | 0.5708 | No | ||
| 55 | ORC2L | 1990470 6510019 | 4085 | 0.025 | 0.5640 | No | ||
| 56 | CDKN1B | 3800025 6450044 | 4101 | 0.024 | 0.5638 | No | ||
| 57 | RBL2 | 580446 1400670 | 4235 | 0.022 | 0.5572 | No | ||
| 58 | CDC25C | 2570673 4760161 6520707 | 4287 | 0.021 | 0.5550 | No | ||
| 59 | PTTG1 | 520463 | 4309 | 0.021 | 0.5544 | No | ||
| 60 | ATM | 3610110 4050524 | 4718 | 0.015 | 0.5328 | No | ||
| 61 | E2F3 | 50162 460180 | 4775 | 0.015 | 0.5302 | No | ||
| 62 | CDKN1A | 4050088 6400706 | 5145 | 0.011 | 0.5105 | No | ||
| 63 | CDC23 | 3190593 | 5170 | 0.011 | 0.5095 | No | ||
| 64 | CHEK1 | 50167 1940300 | 5343 | 0.010 | 0.5005 | No | ||
| 65 | CCNB2 | 6510528 | 5583 | 0.009 | 0.4878 | No | ||
| 66 | YWHAH | 1660133 2810053 | 5727 | 0.008 | 0.4803 | No | ||
| 67 | ANAPC10 | 870086 1170037 2260129 | 6780 | 0.003 | 0.4236 | No | ||
| 68 | CCNA1 | 6290113 | 7824 | 0.000 | 0.3672 | No | ||
| 69 | BUB1 | 5390270 | 8455 | -0.002 | 0.3332 | No | ||
| 70 | CCND2 | 5340167 | 8714 | -0.002 | 0.3194 | No | ||
| 71 | SMC1B | 2450397 | 9457 | -0.004 | 0.2794 | No | ||
| 72 | CDK7 | 2640451 | 10864 | -0.009 | 0.2037 | No | ||
| 73 | CCNB3 | 3360600 | 11501 | -0.013 | 0.1697 | No | ||
| 74 | TGFB2 | 4920292 | 11832 | -0.015 | 0.1523 | No | ||
| 75 | PLK1 | 1780369 2640121 | 13157 | -0.027 | 0.0815 | No | ||
| 76 | CDKN2B | 6020040 | 13499 | -0.032 | 0.0639 | No | ||
| 77 | ANAPC11 | 1780601 | 13517 | -0.032 | 0.0638 | No | ||
| 78 | PRKDC | 1400072 4230541 | 13612 | -0.034 | 0.0596 | No | ||
| 79 | MCM3 | 5570068 | 13752 | -0.036 | 0.0531 | No | ||
| 80 | SFN | 6290301 7510608 | 14188 | -0.045 | 0.0308 | No | ||
| 81 | ANAPC2 | 6660438 | 14206 | -0.046 | 0.0311 | No | ||
| 82 | CDC14B | 2320497 3360162 | 14483 | -0.053 | 0.0176 | No | ||
| 83 | MCM5 | 2680647 | 14988 | -0.071 | -0.0077 | No | ||
| 84 | TP53 | 6130707 | 15076 | -0.075 | -0.0105 | No | ||
| 85 | CDC26 | 5570279 | 15079 | -0.075 | -0.0086 | No | ||
| 86 | CHEK2 | 610139 1050022 | 15148 | -0.079 | -0.0102 | No | ||
| 87 | CDK6 | 4920253 | 15168 | -0.079 | -0.0091 | No | ||
| 88 | CDC20 | 3440017 3440044 6220088 | 15274 | -0.085 | -0.0125 | No | ||
| 89 | GADD45A | 2900717 | 15292 | -0.086 | -0.0111 | No | ||
| 90 | BUB3 | 3170546 | 15296 | -0.087 | -0.0090 | No | ||
| 91 | MCM2 | 5050139 | 15554 | -0.103 | -0.0201 | No | ||
| 92 | CDKN2D | 6040035 | 15762 | -0.118 | -0.0282 | No | ||
| 93 | MAD1L1 | 5700035 | 15806 | -0.122 | -0.0272 | No | ||
| 94 | GADD45B | 2350408 | 15876 | -0.128 | -0.0275 | No | ||
| 95 | TGFB1 | 1940162 | 16050 | -0.146 | -0.0330 | No | ||
| 96 | E2F1 | 5360093 | 16136 | -0.155 | -0.0335 | No | ||
| 97 | CDC25B | 6940102 | 17188 | -0.308 | -0.0821 | No | ||
| 98 | CCNE1 | 110064 | 17434 | -0.356 | -0.0858 | No | ||
| 99 | ANAPC7 | 5260274 | 17762 | -0.450 | -0.0915 | No | ||
| 100 | SMAD3 | 6450671 | 18084 | -0.607 | -0.0927 | No | ||
| 101 | CDKN2C | 5050750 5130148 | 18240 | -0.763 | -0.0808 | No | ||
| 102 | MAD2L2 | 1240358 | 18254 | -0.781 | -0.0607 | No | ||
| 103 | FZR1 | 6220504 6660670 | 18331 | -0.915 | -0.0405 | No | ||
| 104 | CDK4 | 540075 4540600 | 18574 | -2.101 | 0.0023 | No |