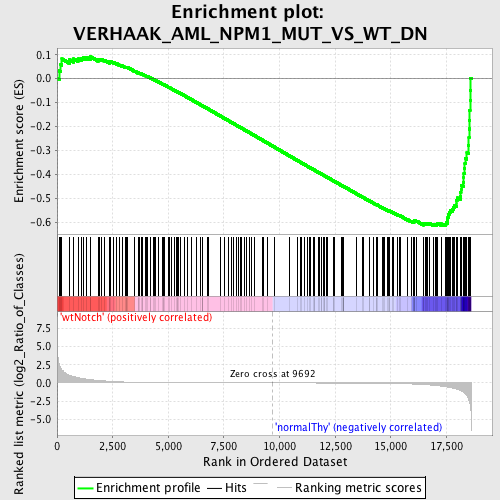
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | VERHAAK_AML_NPM1_MUT_VS_WT_DN |
| Enrichment Score (ES) | -0.6156009 |
| Normalized Enrichment Score (NES) | -1.573249 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.30430895 |
| FWER p-Value | 0.954 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ITM2A | 460576 4210086 | 94 | 2.617 | 0.0335 | No | ||
| 2 | DPYSL2 | 2100427 3130112 5700324 | 166 | 1.977 | 0.0589 | No | ||
| 3 | PALM | 130601 3850017 | 217 | 1.788 | 0.0825 | No | ||
| 4 | DLK1 | 1240138 | 569 | 1.036 | 0.0788 | No | ||
| 5 | HSPG2 | 2510687 6220750 | 743 | 0.893 | 0.0826 | No | ||
| 6 | FLNB | 2510575 | 940 | 0.729 | 0.0827 | No | ||
| 7 | ALDH2 | 4230019 | 1086 | 0.637 | 0.0842 | No | ||
| 8 | H1F0 | 1400408 | 1179 | 0.588 | 0.0879 | No | ||
| 9 | APP | 2510053 | 1312 | 0.534 | 0.0886 | No | ||
| 10 | HSPB1 | 6760435 | 1500 | 0.457 | 0.0852 | No | ||
| 11 | ITM2C | 780647 | 1512 | 0.452 | 0.0913 | No | ||
| 12 | LMO4 | 3800746 | 1841 | 0.351 | 0.0787 | No | ||
| 13 | AK3L1 | 7100601 | 1901 | 0.335 | 0.0804 | No | ||
| 14 | ASS1 | 1580632 4070082 | 1996 | 0.311 | 0.0799 | No | ||
| 15 | SLC38A1 | 520465 | 2110 | 0.281 | 0.0779 | No | ||
| 16 | POLE | 6020538 | 2348 | 0.226 | 0.0684 | No | ||
| 17 | ATP9A | 4780673 5390280 | 2359 | 0.225 | 0.0712 | No | ||
| 18 | MX1 | 2690301 6840364 | 2414 | 0.215 | 0.0714 | No | ||
| 19 | TGFBI | 2060446 6900112 | 2530 | 0.191 | 0.0680 | No | ||
| 20 | SERPING1 | 5550440 | 2664 | 0.171 | 0.0633 | No | ||
| 21 | CHI3L1 | 1170039 | 2817 | 0.150 | 0.0573 | No | ||
| 22 | WASF1 | 2060484 | 2925 | 0.136 | 0.0535 | No | ||
| 23 | RPS6KA2 | 2970161 4730278 | 3077 | 0.123 | 0.0471 | No | ||
| 24 | RUNX3 | 3800546 | 3117 | 0.119 | 0.0468 | No | ||
| 25 | PPP1R16B | 3520300 | 3138 | 0.117 | 0.0474 | No | ||
| 26 | TCF3 | 5080022 | 3162 | 0.115 | 0.0478 | No | ||
| 27 | ZAP70 | 1410494 2260504 | 3498 | 0.085 | 0.0309 | No | ||
| 28 | GNG7 | 1940019 | 3651 | 0.074 | 0.0238 | No | ||
| 29 | GYPC | 2320102 | 3658 | 0.073 | 0.0245 | No | ||
| 30 | PRKD3 | 5220520 5890519 | 3719 | 0.069 | 0.0223 | No | ||
| 31 | ELA2 | 1570452 5270129 | 3772 | 0.066 | 0.0204 | No | ||
| 32 | SERPINF1 | 7040367 | 3851 | 0.061 | 0.0171 | No | ||
| 33 | HIST1H2BG | 5670632 | 3859 | 0.061 | 0.0176 | No | ||
| 34 | ERG | 50154 1770739 | 3953 | 0.057 | 0.0134 | No | ||
| 35 | NEDD4 | 460736 1190280 | 4015 | 0.053 | 0.0109 | No | ||
| 36 | TNFRSF21 | 6380100 | 4046 | 0.052 | 0.0100 | No | ||
| 37 | CAV1 | 870025 | 4187 | 0.047 | 0.0031 | No | ||
| 38 | MSLN | 2640048 | 4195 | 0.046 | 0.0034 | No | ||
| 39 | GALC | 2260685 4810161 6370040 | 4354 | 0.041 | -0.0045 | No | ||
| 40 | GSN | 3830168 | 4386 | 0.041 | -0.0056 | No | ||
| 41 | CD69 | 380167 4730088 | 4401 | 0.040 | -0.0058 | No | ||
| 42 | FZD2 | 6480154 | 4543 | 0.037 | -0.0129 | No | ||
| 43 | OPTN | 3940041 | 4734 | 0.032 | -0.0227 | No | ||
| 44 | SERPINE2 | 2810390 | 4785 | 0.031 | -0.0250 | No | ||
| 45 | PDE3B | 2030563 | 4803 | 0.031 | -0.0254 | No | ||
| 46 | B4GALT6 | 1850176 2340687 | 4827 | 0.031 | -0.0262 | No | ||
| 47 | MPL | 2360253 | 5021 | 0.027 | -0.0363 | No | ||
| 48 | NBL1 | 2480021 | 5034 | 0.027 | -0.0365 | No | ||
| 49 | CSF3R | 670500 2640239 3440091 5690020 | 5120 | 0.026 | -0.0408 | No | ||
| 50 | CD48 | 1570647 | 5258 | 0.024 | -0.0479 | No | ||
| 51 | CCR2 | 1450519 | 5353 | 0.023 | -0.0526 | No | ||
| 52 | GATM | 380671 | 5417 | 0.022 | -0.0557 | No | ||
| 53 | SH3BP4 | 3140242 | 5432 | 0.022 | -0.0562 | No | ||
| 54 | RBPMS | 3990494 | 5451 | 0.022 | -0.0568 | No | ||
| 55 | FCER1A | 1050671 | 5560 | 0.020 | -0.0624 | No | ||
| 56 | CD34 | 6650270 | 5733 | 0.019 | -0.0714 | No | ||
| 57 | FGFR1 | 2190095 4570161 | 5874 | 0.017 | -0.0788 | No | ||
| 58 | KCNE1L | 4670092 | 6052 | 0.016 | -0.0881 | No | ||
| 59 | PTPRM | 6370136 | 6248 | 0.014 | -0.0985 | No | ||
| 60 | SYNGR1 | 5130452 6650056 | 6434 | 0.013 | -0.1084 | No | ||
| 61 | TM6SF1 | 730181 | 6528 | 0.012 | -0.1132 | No | ||
| 62 | SPARC | 1690086 | 6541 | 0.012 | -0.1137 | No | ||
| 63 | PTGER4 | 6400670 | 6552 | 0.012 | -0.1140 | No | ||
| 64 | ABLIM1 | 580170 3710338 6520504 | 6741 | 0.011 | -0.1241 | No | ||
| 65 | HGF | 3360593 | 6813 | 0.011 | -0.1278 | No | ||
| 66 | P2RY5 | 3800136 | 7353 | 0.008 | -0.1569 | No | ||
| 67 | MYH11 | 7100273 | 7506 | 0.008 | -0.1650 | No | ||
| 68 | MEST | 4780440 6620292 | 7694 | 0.007 | -0.1751 | No | ||
| 69 | GPA33 | 2650044 | 7842 | 0.006 | -0.1830 | No | ||
| 70 | ARHGEF12 | 3990195 | 7948 | 0.006 | -0.1886 | No | ||
| 71 | MST1 | 1400403 | 8042 | 0.005 | -0.1935 | No | ||
| 72 | ARHGEF17 | 2060008 | 8134 | 0.005 | -0.1984 | No | ||
| 73 | CLEC5A | 6420139 | 8222 | 0.005 | -0.2031 | No | ||
| 74 | EPS8 | 7050204 | 8285 | 0.005 | -0.2064 | No | ||
| 75 | NPR3 | 1580239 | 8419 | 0.004 | -0.2135 | No | ||
| 76 | STK32B | 7000204 | 8532 | 0.004 | -0.2195 | No | ||
| 77 | ST18 | 5720537 | 8669 | 0.003 | -0.2269 | No | ||
| 78 | FZD6 | 360288 | 8729 | 0.003 | -0.2300 | No | ||
| 79 | PTRF | 2260204 | 8890 | 0.002 | -0.2387 | No | ||
| 80 | TRH | 5050576 | 9211 | 0.001 | -0.2560 | No | ||
| 81 | GNAI1 | 4560390 | 9272 | 0.001 | -0.2592 | No | ||
| 82 | EVI1 | 2000193 2680494 | 9476 | 0.001 | -0.2702 | No | ||
| 83 | IFI16 | 6520601 | 9777 | -0.000 | -0.2865 | No | ||
| 84 | PPP4R2 | 5890035 | 10436 | -0.002 | -0.3222 | No | ||
| 85 | CD19 | 6940692 | 10822 | -0.003 | -0.3430 | No | ||
| 86 | GPM6B | 6220458 | 10936 | -0.004 | -0.3491 | No | ||
| 87 | ICAM4 | 6770053 | 10978 | -0.004 | -0.3513 | No | ||
| 88 | BANK1 | 50750 | 11124 | -0.004 | -0.3591 | No | ||
| 89 | CD38 | 6100056 | 11248 | -0.005 | -0.3657 | No | ||
| 90 | BAALC | 5310133 6450170 | 11324 | -0.005 | -0.3697 | No | ||
| 91 | NT5E | 2030603 1400731 | 11378 | -0.005 | -0.3725 | No | ||
| 92 | PAM | 5290528 | 11508 | -0.006 | -0.3794 | No | ||
| 93 | VLDLR | 870722 3060047 5340452 6550131 | 11548 | -0.006 | -0.3814 | No | ||
| 94 | DACH1 | 2450593 5700592 | 11569 | -0.006 | -0.3824 | No | ||
| 95 | DCHS1 | 4850487 | 11756 | -0.007 | -0.3924 | No | ||
| 96 | TRPM4 | 6980270 | 11803 | -0.007 | -0.3948 | No | ||
| 97 | SPON1 | 1660301 | 11887 | -0.007 | -0.3992 | No | ||
| 98 | SCRN1 | 6040025 6580019 | 11896 | -0.007 | -0.3995 | No | ||
| 99 | GPSM2 | 2260053 | 11965 | -0.008 | -0.4031 | No | ||
| 100 | TMOD1 | 3850100 | 12014 | -0.008 | -0.4056 | No | ||
| 101 | P2RX5 | 5570427 6130435 | 12021 | -0.008 | -0.4058 | No | ||
| 102 | CLDN15 | 4150270 4730592 | 12118 | -0.008 | -0.4109 | No | ||
| 103 | MLLT3 | 510204 3850722 6200088 | 12161 | -0.009 | -0.4130 | No | ||
| 104 | MEF2C | 670025 780338 | 12418 | -0.010 | -0.4267 | No | ||
| 105 | TKTL1 | 2810672 6760102 | 12426 | -0.010 | -0.4270 | No | ||
| 106 | TRAF5 | 3290064 | 12470 | -0.010 | -0.4292 | No | ||
| 107 | DHRS3 | 360609 | 12770 | -0.012 | -0.4452 | No | ||
| 108 | PROM1 | 3170537 | 12810 | -0.012 | -0.4471 | No | ||
| 109 | ROBO1 | 3710398 | 12858 | -0.013 | -0.4495 | No | ||
| 110 | PMAIP1 | 1400717 5360047 5900039 | 12864 | -0.013 | -0.4496 | No | ||
| 111 | POU4F1 | 2260315 | 12873 | -0.013 | -0.4498 | No | ||
| 112 | BLNK | 70288 6590092 | 12874 | -0.013 | -0.4497 | No | ||
| 113 | AGTPBP1 | 1990735 | 13437 | -0.017 | -0.4799 | No | ||
| 114 | IL5RA | 4540091 | 13732 | -0.021 | -0.4955 | No | ||
| 115 | PDE4B | 4480121 | 13783 | -0.022 | -0.4979 | No | ||
| 116 | RETN | 1050041 | 14024 | -0.026 | -0.5106 | No | ||
| 117 | TGFB1I1 | 2060288 6550450 | 14048 | -0.026 | -0.5114 | No | ||
| 118 | TSPAN7 | 870133 | 14234 | -0.030 | -0.5210 | No | ||
| 119 | CYP2E1 | 4480209 | 14353 | -0.032 | -0.5270 | No | ||
| 120 | NCAM1 | 3140026 | 14373 | -0.033 | -0.5275 | No | ||
| 121 | CST7 | 380369 | 14381 | -0.033 | -0.5274 | No | ||
| 122 | ADCY3 | 2350347 3440242 | 14619 | -0.039 | -0.5397 | No | ||
| 123 | MS4A3 | 2480309 2690086 | 14656 | -0.040 | -0.5411 | No | ||
| 124 | SPRY1 | 450253 460093 | 14725 | -0.043 | -0.5441 | No | ||
| 125 | RGS1 | 4060347 4540181 | 14872 | -0.048 | -0.5513 | No | ||
| 126 | ZHX2 | 2900452 | 14917 | -0.051 | -0.5530 | No | ||
| 127 | RHOBTB3 | 4010687 | 14925 | -0.051 | -0.5526 | No | ||
| 128 | F2RL1 | 3290279 | 14955 | -0.053 | -0.5534 | No | ||
| 129 | EGFL7 | 150113 670170 3840707 | 15073 | -0.058 | -0.5589 | No | ||
| 130 | TSPAN13 | 1690450 | 15135 | -0.062 | -0.5613 | No | ||
| 131 | FHL1 | 60278 | 15297 | -0.073 | -0.5689 | No | ||
| 132 | TFPI | 1400288 5550008 | 15382 | -0.079 | -0.5723 | No | ||
| 133 | BPGM | 5080520 | 15423 | -0.083 | -0.5733 | No | ||
| 134 | LHFPL2 | 450020 | 15765 | -0.117 | -0.5900 | No | ||
| 135 | MEF2A | 2350041 3360128 6040180 | 15910 | -0.135 | -0.5959 | No | ||
| 136 | CACNA2D2 | 520736 | 16007 | -0.149 | -0.5989 | No | ||
| 137 | CD200 | 940725 5260647 | 16036 | -0.151 | -0.5982 | No | ||
| 138 | CD7 | 3140465 | 16054 | -0.155 | -0.5968 | No | ||
| 139 | HPGD | 6770192 | 16057 | -0.155 | -0.5946 | No | ||
| 140 | DEPDC6 | 730326 5820239 | 16066 | -0.157 | -0.5927 | No | ||
| 141 | CD2 | 430672 | 16174 | -0.174 | -0.5960 | No | ||
| 142 | TACSTD2 | 6660341 | 16465 | -0.223 | -0.6084 | No | ||
| 143 | SNCA | 5340673 | 16479 | -0.225 | -0.6058 | No | ||
| 144 | IGFBP7 | 520411 3060110 5290152 | 16558 | -0.240 | -0.6065 | No | ||
| 145 | CIITA | 2650242 | 16605 | -0.249 | -0.6053 | No | ||
| 146 | NARFL | 2260372 4050341 | 16649 | -0.255 | -0.6039 | No | ||
| 147 | KIT | 7040095 | 16733 | -0.270 | -0.6044 | No | ||
| 148 | PTGIR | 2100563 | 16902 | -0.313 | -0.6089 | Yes | ||
| 149 | YPEL1 | 6590435 | 16994 | -0.337 | -0.6088 | Yes | ||
| 150 | PTPRCAP | 6650075 | 17074 | -0.364 | -0.6078 | Yes | ||
| 151 | CENTD1 | 2570592 | 17107 | -0.376 | -0.6039 | Yes | ||
| 152 | BZRAP1 | 5290487 | 17289 | -0.448 | -0.6071 | Yes | ||
| 153 | ITGA6 | 3830129 | 17446 | -0.521 | -0.6079 | Yes | ||
| 154 | RAMP1 | 2320168 | 17482 | -0.534 | -0.6019 | Yes | ||
| 155 | JUP | 2510671 | 17545 | -0.566 | -0.5969 | Yes | ||
| 156 | PRTN3 | 6900670 | 17554 | -0.570 | -0.5890 | Yes | ||
| 157 | VPREB1 | 3520280 | 17565 | -0.574 | -0.5810 | Yes | ||
| 158 | GATA3 | 6130068 | 17582 | -0.581 | -0.5733 | Yes | ||
| 159 | ADIPOR1 | 3140440 | 17600 | -0.588 | -0.5656 | Yes | ||
| 160 | RRAGD | 670075 | 17623 | -0.601 | -0.5579 | Yes | ||
| 161 | CD3D | 2810739 | 17668 | -0.628 | -0.5510 | Yes | ||
| 162 | MMP9 | 580338 | 17760 | -0.688 | -0.5458 | Yes | ||
| 163 | GJA1 | 5220731 | 17809 | -0.725 | -0.5377 | Yes | ||
| 164 | FOXO1A | 3120670 | 17857 | -0.757 | -0.5291 | Yes | ||
| 165 | VIL2 | 5570161 | 17954 | -0.833 | -0.5220 | Yes | ||
| 166 | ISG20 | 1770750 | 17956 | -0.834 | -0.5097 | Yes | ||
| 167 | EVL | 1740113 | 17980 | -0.850 | -0.4984 | Yes | ||
| 168 | TRIB2 | 4120605 | 18126 | -1.035 | -0.4910 | Yes | ||
| 169 | PLXND1 | 5270682 | 18141 | -1.047 | -0.4763 | Yes | ||
| 170 | UGCG | 3420053 | 18155 | -1.067 | -0.4613 | Yes | ||
| 171 | LCN2 | 2510112 | 18163 | -1.085 | -0.4456 | Yes | ||
| 172 | LAMC1 | 4730025 | 18255 | -1.255 | -0.4320 | Yes | ||
| 173 | GMPR | 3990022 | 18280 | -1.324 | -0.4138 | Yes | ||
| 174 | EHD3 | 5360484 | 18286 | -1.339 | -0.3943 | Yes | ||
| 175 | SIDT1 | 7100010 | 18319 | -1.420 | -0.3751 | Yes | ||
| 176 | SORL1 | 630204 | 18331 | -1.454 | -0.3542 | Yes | ||
| 177 | MPO | 2360176 2760440 5690176 | 18349 | -1.516 | -0.3327 | Yes | ||
| 178 | IFITM1 | 1690601 3120438 6510075 | 18422 | -1.788 | -0.3103 | Yes | ||
| 179 | EMP1 | 4120438 | 18497 | -2.284 | -0.2806 | Yes | ||
| 180 | ALAS2 | 6550176 | 18511 | -2.385 | -0.2461 | Yes | ||
| 181 | DNTT | 670162 | 18519 | -2.472 | -0.2100 | Yes | ||
| 182 | FGF13 | 630575 1570440 5360121 | 18534 | -2.575 | -0.1727 | Yes | ||
| 183 | CCND2 | 5340167 | 18548 | -2.713 | -0.1334 | Yes | ||
| 184 | SSBP2 | 2260725 2900400 4730711 | 18561 | -2.849 | -0.0920 | Yes | ||
| 185 | TOX | 3440053 | 18571 | -3.029 | -0.0478 | Yes | ||
| 186 | PRODH | 1240452 | 18588 | -3.398 | 0.0015 | Yes |

