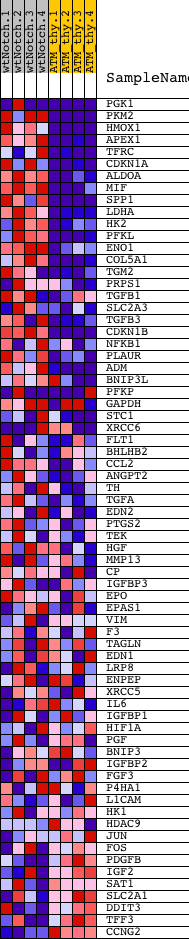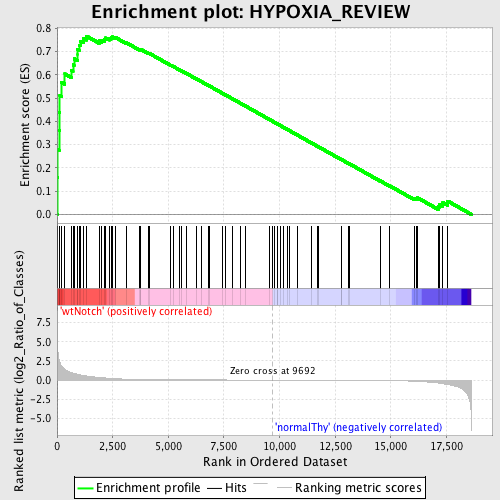
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | wtNotch |
| GeneSet | HYPOXIA_REVIEW |
| Enrichment Score (ES) | 0.7661528 |
| Normalized Enrichment Score (NES) | 1.6750206 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.024416596 |
| FWER p-Value | 0.16 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PGK1 | 1570494 630300 | 8 | 5.004 | 0.1594 | Yes | ||
| 2 | PKM2 | 6520403 70500 | 34 | 3.793 | 0.2792 | Yes | ||
| 3 | HMOX1 | 1740687 | 92 | 2.622 | 0.3599 | Yes | ||
| 4 | APEX1 | 3190519 | 108 | 2.501 | 0.4390 | Yes | ||
| 5 | TFRC | 4050551 | 123 | 2.289 | 0.5113 | Yes | ||
| 6 | CDKN1A | 4050088 6400706 | 188 | 1.874 | 0.5677 | Yes | ||
| 7 | ALDOA | 6290672 | 351 | 1.397 | 0.6036 | Yes | ||
| 8 | MIF | 3710332 | 650 | 0.957 | 0.6181 | Yes | ||
| 9 | SPP1 | 2470609 | 716 | 0.913 | 0.6437 | Yes | ||
| 10 | LDHA | 2190594 | 764 | 0.880 | 0.6693 | Yes | ||
| 11 | HK2 | 2640722 | 915 | 0.752 | 0.6852 | Yes | ||
| 12 | PFKL | 6200167 | 918 | 0.751 | 0.7091 | Yes | ||
| 13 | ENO1 | 5340128 | 992 | 0.689 | 0.7272 | Yes | ||
| 14 | COL5A1 | 2230050 | 1050 | 0.656 | 0.7451 | Yes | ||
| 15 | TGM2 | 5360452 | 1189 | 0.585 | 0.7563 | Yes | ||
| 16 | PRPS1 | 6350129 | 1321 | 0.529 | 0.7662 | Yes | ||
| 17 | TGFB1 | 1940162 | 1890 | 0.338 | 0.7463 | No | ||
| 18 | SLC2A3 | 1990377 | 2016 | 0.304 | 0.7493 | No | ||
| 19 | TGFB3 | 1070041 | 2137 | 0.274 | 0.7516 | No | ||
| 20 | CDKN1B | 3800025 6450044 | 2173 | 0.267 | 0.7582 | No | ||
| 21 | NFKB1 | 5420358 | 2339 | 0.229 | 0.7567 | No | ||
| 22 | PLAUR | 5910280 | 2423 | 0.213 | 0.7590 | No | ||
| 23 | ADM | 4810121 | 2472 | 0.202 | 0.7628 | No | ||
| 24 | BNIP3L | 1940347 | 2638 | 0.174 | 0.7595 | No | ||
| 25 | PFKP | 70138 6760040 1170278 | 3109 | 0.120 | 0.7380 | No | ||
| 26 | GAPDH | 110022 430039 6220161 | 3708 | 0.070 | 0.7080 | No | ||
| 27 | STC1 | 360161 | 3750 | 0.067 | 0.7079 | No | ||
| 28 | XRCC6 | 2850450 3870528 | 3758 | 0.066 | 0.7097 | No | ||
| 29 | FLT1 | 3830167 4920438 | 4096 | 0.050 | 0.6931 | No | ||
| 30 | BHLHB2 | 7040603 | 4158 | 0.048 | 0.6913 | No | ||
| 31 | CCL2 | 4760019 | 5118 | 0.026 | 0.6404 | No | ||
| 32 | ANGPT2 | 1230458 5720706 6040605 | 5242 | 0.024 | 0.6346 | No | ||
| 33 | TH | 2100056 | 5511 | 0.021 | 0.6208 | No | ||
| 34 | TGFA | 1570332 | 5600 | 0.020 | 0.6167 | No | ||
| 35 | EDN2 | 6760647 | 5815 | 0.018 | 0.6057 | No | ||
| 36 | PTGS2 | 2510301 3170369 | 6277 | 0.014 | 0.5813 | No | ||
| 37 | TEK | 6350541 | 6505 | 0.012 | 0.5695 | No | ||
| 38 | HGF | 3360593 | 6813 | 0.011 | 0.5532 | No | ||
| 39 | MMP13 | 510471 3940097 | 6828 | 0.011 | 0.5528 | No | ||
| 40 | CP | 2570484 | 7415 | 0.008 | 0.5215 | No | ||
| 41 | IGFBP3 | 2370500 | 7579 | 0.007 | 0.5129 | No | ||
| 42 | EPO | 940180 | 7898 | 0.006 | 0.4960 | No | ||
| 43 | EPAS1 | 5290156 | 8247 | 0.005 | 0.4774 | No | ||
| 44 | VIM | 20431 | 8480 | 0.004 | 0.4650 | No | ||
| 45 | F3 | 2940180 | 9553 | 0.000 | 0.4072 | No | ||
| 46 | TAGLN | 6660280 | 9658 | 0.000 | 0.4016 | No | ||
| 47 | EDN1 | 1770047 | 9775 | -0.000 | 0.3953 | No | ||
| 48 | LRP8 | 3610746 5360035 | 9894 | -0.001 | 0.3890 | No | ||
| 49 | ENPEP | 130373 | 9918 | -0.001 | 0.3878 | No | ||
| 50 | XRCC5 | 7100286 | 10048 | -0.001 | 0.3808 | No | ||
| 51 | IL6 | 380133 | 10180 | -0.001 | 0.3738 | No | ||
| 52 | IGFBP1 | 5720435 | 10359 | -0.002 | 0.3643 | No | ||
| 53 | HIF1A | 5670605 | 10440 | -0.002 | 0.3601 | No | ||
| 54 | PGF | 4810593 | 10786 | -0.003 | 0.3416 | No | ||
| 55 | BNIP3 | 3140270 | 11417 | -0.006 | 0.3078 | No | ||
| 56 | IGFBP2 | 5220563 | 11715 | -0.007 | 0.2920 | No | ||
| 57 | FGF3 | 4230685 | 11751 | -0.007 | 0.2903 | No | ||
| 58 | P4HA1 | 2630692 | 12769 | -0.012 | 0.2358 | No | ||
| 59 | L1CAM | 4850021 | 12783 | -0.012 | 0.2355 | No | ||
| 60 | HK1 | 4280402 | 13116 | -0.015 | 0.2181 | No | ||
| 61 | HDAC9 | 1990010 2260133 | 13130 | -0.015 | 0.2179 | No | ||
| 62 | JUN | 840170 | 14517 | -0.036 | 0.1443 | No | ||
| 63 | FOS | 1850315 | 14961 | -0.053 | 0.1221 | No | ||
| 64 | PDGFB | 3060440 6370008 | 16044 | -0.153 | 0.0686 | No | ||
| 65 | IGF2 | 6510020 | 16150 | -0.170 | 0.0684 | No | ||
| 66 | SAT1 | 4570463 | 16198 | -0.177 | 0.0715 | No | ||
| 67 | SLC2A1 | 2100609 | 17145 | -0.391 | 0.0330 | No | ||
| 68 | DDIT3 | 780373 | 17175 | -0.400 | 0.0442 | No | ||
| 69 | TFF3 | 1580129 | 17339 | -0.476 | 0.0506 | No | ||
| 70 | CCNG2 | 3190095 | 17555 | -0.570 | 0.0572 | No |