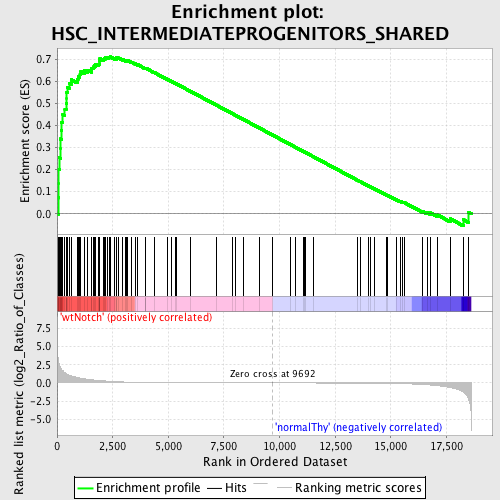
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | wtNotch |
| GeneSet | HSC_INTERMEDIATEPROGENITORS_SHARED |
| Enrichment Score (ES) | 0.7110721 |
| Normalized Enrichment Score (NES) | 1.6051704 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.083871074 |
| FWER p-Value | 0.672 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HES6 | 540411 6550504 | 45 | 3.458 | 0.0719 | Yes | ||
| 2 | BID | 4590592 6130008 | 56 | 3.112 | 0.1382 | Yes | ||
| 3 | SDC4 | 6370411 | 61 | 3.005 | 0.2025 | Yes | ||
| 4 | APEX1 | 3190519 | 108 | 2.501 | 0.2538 | Yes | ||
| 5 | C12ORF10 | 4120324 5270113 | 152 | 2.053 | 0.2956 | Yes | ||
| 6 | KLHDC4 | 870288 5900092 | 157 | 2.036 | 0.3391 | Yes | ||
| 7 | PTCD2 | 1230088 1400605 3170576 6200609 | 192 | 1.857 | 0.3771 | Yes | ||
| 8 | MAPRE2 | 1660435 5860403 | 207 | 1.835 | 0.4158 | Yes | ||
| 9 | NPC2 | 2350102 | 257 | 1.680 | 0.4493 | Yes | ||
| 10 | ALDOA | 6290672 | 351 | 1.397 | 0.4742 | Yes | ||
| 11 | GALK1 | 840162 | 406 | 1.288 | 0.4990 | Yes | ||
| 12 | TBRG4 | 3610239 6370594 | 432 | 1.220 | 0.5239 | Yes | ||
| 13 | SDF2L1 | 4230446 | 435 | 1.218 | 0.5499 | Yes | ||
| 14 | DDX52 | 2970411 | 474 | 1.157 | 0.5728 | Yes | ||
| 15 | GTF3A | 2340451 | 545 | 1.059 | 0.5917 | Yes | ||
| 16 | CHCHD4 | 2680075 | 645 | 0.960 | 0.6070 | Yes | ||
| 17 | MRPL12 | 3120112 | 907 | 0.760 | 0.6093 | Yes | ||
| 18 | DBI | 1090706 | 943 | 0.726 | 0.6230 | Yes | ||
| 19 | APIP | 2480440 2810471 | 1027 | 0.667 | 0.6328 | Yes | ||
| 20 | ITGB4BP | 1400541 | 1067 | 0.647 | 0.6446 | Yes | ||
| 21 | PLAC8 | 2900019 | 1228 | 0.565 | 0.6481 | Yes | ||
| 22 | TEX2 | 1740504 2480102 3060494 | 1373 | 0.508 | 0.6513 | Yes | ||
| 23 | WDR34 | 6100215 | 1549 | 0.438 | 0.6512 | Yes | ||
| 24 | COMMD10 | 3120451 | 1552 | 0.437 | 0.6605 | Yes | ||
| 25 | GFM1 | 6220711 | 1629 | 0.413 | 0.6653 | Yes | ||
| 26 | EEF1B2 | 4850400 6130161 | 1666 | 0.402 | 0.6720 | Yes | ||
| 27 | PES1 | 2470427 | 1731 | 0.382 | 0.6767 | Yes | ||
| 28 | POLR3E | 380152 | 1855 | 0.347 | 0.6776 | Yes | ||
| 29 | THOC5 | 4570047 | 1893 | 0.337 | 0.6828 | Yes | ||
| 30 | MRPS35 | 2370603 4070551 | 1896 | 0.336 | 0.6899 | Yes | ||
| 31 | GPT2 | 1850463 | 1898 | 0.336 | 0.6971 | Yes | ||
| 32 | SSR2 | 5390180 | 1923 | 0.328 | 0.7028 | Yes | ||
| 33 | MRPS25 | 610441 870128 1410368 | 2101 | 0.283 | 0.6993 | Yes | ||
| 34 | DDX20 | 3290348 | 2139 | 0.274 | 0.7032 | Yes | ||
| 35 | GMPPA | 520450 3120497 | 2178 | 0.267 | 0.7069 | Yes | ||
| 36 | LMAN2 | 430154 | 2251 | 0.249 | 0.7084 | Yes | ||
| 37 | CTSZ | 1500687 1690364 | 2333 | 0.230 | 0.7089 | Yes | ||
| 38 | SURF6 | 3060451 | 2382 | 0.220 | 0.7111 | Yes | ||
| 39 | TXNDC4 | 450537 | 2590 | 0.182 | 0.7038 | No | ||
| 40 | RSU1 | 4850372 | 2668 | 0.170 | 0.7033 | No | ||
| 41 | CFDP1 | 7000112 | 2679 | 0.169 | 0.7064 | No | ||
| 42 | DHX37 | 1400309 | 2748 | 0.159 | 0.7061 | No | ||
| 43 | TCN2 | 2810139 | 2931 | 0.136 | 0.6992 | No | ||
| 44 | PCCA | 3390400 | 3076 | 0.123 | 0.6941 | No | ||
| 45 | PFKP | 70138 6760040 1170278 | 3109 | 0.120 | 0.6950 | No | ||
| 46 | CPA3 | 610242 6380102 | 3170 | 0.114 | 0.6942 | No | ||
| 47 | MAPKAPK3 | 630520 | 3350 | 0.097 | 0.6866 | No | ||
| 48 | POLR3C | 6940546 | 3500 | 0.085 | 0.6804 | No | ||
| 49 | GCNT3 | 6290239 | 3627 | 0.076 | 0.6752 | No | ||
| 50 | TNS3 | 6380242 | 3966 | 0.056 | 0.6582 | No | ||
| 51 | AP3S2 | 2630411 5080053 | 3970 | 0.056 | 0.6592 | No | ||
| 52 | CDADC1 | 3710201 4120064 | 3990 | 0.055 | 0.6594 | No | ||
| 53 | SIGLEC5 | 5690398 | 4364 | 0.041 | 0.6401 | No | ||
| 54 | CEBPZ | 2970053 2680068 | 4980 | 0.028 | 0.6075 | No | ||
| 55 | MDH1 | 6660358 6760731 | 5146 | 0.025 | 0.5992 | No | ||
| 56 | C5ORF14 | 3780050 | 5306 | 0.023 | 0.5911 | No | ||
| 57 | MAOA | 1410039 4610324 | 5355 | 0.023 | 0.5890 | No | ||
| 58 | ATP6AP2 | 4010059 5080315 7100347 6660484 | 5991 | 0.016 | 0.5551 | No | ||
| 59 | GPR180 | 5390711 | 7153 | 0.009 | 0.4926 | No | ||
| 60 | DTX4 | 4120433 | 7888 | 0.006 | 0.4531 | No | ||
| 61 | PLAA | 60706 6370369 | 8028 | 0.006 | 0.4457 | No | ||
| 62 | NEBL | 1850369 2480286 | 8356 | 0.004 | 0.4282 | No | ||
| 63 | CITED2 | 5670114 5130088 | 9116 | 0.002 | 0.3873 | No | ||
| 64 | RPN1 | 1240113 | 9669 | 0.000 | 0.3575 | No | ||
| 65 | ELOVL3 | 6860050 | 10476 | -0.002 | 0.3140 | No | ||
| 66 | AS3MT | 1570064 2350113 | 10731 | -0.003 | 0.3004 | No | ||
| 67 | NAT12 | 1780092 4730370 | 11071 | -0.004 | 0.2822 | No | ||
| 68 | GYS1 | 540154 | 11098 | -0.004 | 0.2809 | No | ||
| 69 | BET1 | 4010725 | 11151 | -0.005 | 0.2782 | No | ||
| 70 | AHCY | 4230605 | 11511 | -0.006 | 0.2589 | No | ||
| 71 | STATIP1 | 4480022 6940097 | 13490 | -0.018 | 0.1526 | No | ||
| 72 | CUL5 | 450142 | 13630 | -0.020 | 0.1455 | No | ||
| 73 | NIPSNAP3B | 630672 3710091 4540288 4780576 | 13993 | -0.025 | 0.1265 | No | ||
| 74 | D4ST1 | 1780671 | 14071 | -0.026 | 0.1229 | No | ||
| 75 | HDGF | 5130114 5420066 6590152 | 14274 | -0.030 | 0.1126 | No | ||
| 76 | SNUPN | 5050131 | 14815 | -0.046 | 0.0845 | No | ||
| 77 | ATP6V1F | 5420082 | 14831 | -0.047 | 0.0847 | No | ||
| 78 | RASSF4 | 510372 1660673 6290040 | 15257 | -0.070 | 0.0632 | No | ||
| 79 | PAPSS2 | 870113 | 15439 | -0.085 | 0.0553 | No | ||
| 80 | SFXN4 | 60750 4560059 | 15541 | -0.094 | 0.0519 | No | ||
| 81 | NUP37 | 2370097 6370435 6380008 | 15598 | -0.099 | 0.0510 | No | ||
| 82 | DYNLL1 | 5570113 | 16427 | -0.216 | 0.0109 | No | ||
| 83 | SCARB1 | 510441 6770047 | 16633 | -0.253 | 0.0053 | No | ||
| 84 | BLOC1S2 | 1240215 | 16771 | -0.278 | 0.0039 | No | ||
| 85 | ERCC8 | 1240300 6450372 6590180 | 17095 | -0.372 | -0.0056 | No | ||
| 86 | PYCARD | 2100750 | 17672 | -0.629 | -0.0231 | No | ||
| 87 | SCIN | 5420180 | 18271 | -1.309 | -0.0273 | No | ||
| 88 | CASP7 | 4210156 | 18481 | -2.135 | 0.0073 | No |

