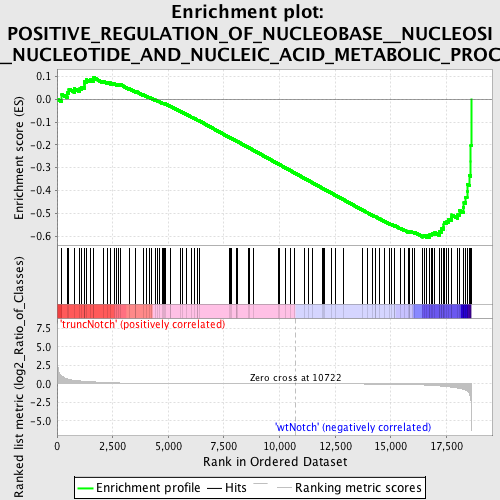
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | POSITIVE_REGULATION_OF_NUCLEOBASE__NUCLEOSIDE__NUCLEOTIDE_AND_NUCLEIC_ACID_METABOLIC_PROCESS |
| Enrichment Score (ES) | -0.60596603 |
| Normalized Enrichment Score (NES) | -1.5770233 |
| Nominal p-value | 0.0027322404 |
| FDR q-value | 0.23398727 |
| FWER p-Value | 0.974 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | EGR1 | 4610347 | 214 | 1.010 | 0.0207 | No | ||
| 2 | ERCC2 | 2360750 4060390 6550138 | 470 | 0.657 | 0.0278 | No | ||
| 3 | SMAD3 | 6450671 | 532 | 0.607 | 0.0439 | No | ||
| 4 | TNFSF13 | 2650170 | 776 | 0.479 | 0.0461 | No | ||
| 5 | SQSTM1 | 6550056 | 1025 | 0.393 | 0.0452 | No | ||
| 6 | BMP7 | 870039 | 1090 | 0.375 | 0.0537 | No | ||
| 7 | HMGA1 | 6580408 | 1226 | 0.349 | 0.0576 | No | ||
| 8 | SCAP | 3800706 | 1230 | 0.349 | 0.0685 | No | ||
| 9 | SMARCD1 | 3060193 3850184 6400369 | 1240 | 0.347 | 0.0791 | No | ||
| 10 | SUPT4H1 | 6200056 | 1313 | 0.332 | 0.0858 | No | ||
| 11 | RBM14 | 5340731 5690315 | 1486 | 0.297 | 0.0860 | No | ||
| 12 | MRPL12 | 3120112 | 1636 | 0.270 | 0.0866 | No | ||
| 13 | NUP62 | 1240128 | 1640 | 0.269 | 0.0950 | No | ||
| 14 | EPC1 | 1570050 5290095 6900193 | 2078 | 0.205 | 0.0779 | No | ||
| 15 | MYST3 | 5270500 | 2270 | 0.181 | 0.0734 | No | ||
| 16 | TRIM28 | 380292 | 2399 | 0.165 | 0.0717 | No | ||
| 17 | TGFB1 | 1940162 | 2566 | 0.146 | 0.0674 | No | ||
| 18 | SUPT5H | 4150546 | 2670 | 0.135 | 0.0661 | No | ||
| 19 | SPI1 | 1410397 | 2778 | 0.125 | 0.0644 | No | ||
| 20 | MYST1 | 2190397 | 2865 | 0.117 | 0.0635 | No | ||
| 21 | EPAS1 | 5290156 | 3250 | 0.090 | 0.0456 | No | ||
| 22 | DYRK1B | 5860706 | 3519 | 0.076 | 0.0335 | No | ||
| 23 | TP53 | 6130707 | 3540 | 0.075 | 0.0348 | No | ||
| 24 | CAMKK2 | 5270047 | 3860 | 0.062 | 0.0196 | No | ||
| 25 | SMARCD3 | 4070497 | 3998 | 0.058 | 0.0140 | No | ||
| 26 | HNF4A | 6620440 | 4142 | 0.053 | 0.0080 | No | ||
| 27 | ELL3 | 1770484 | 4241 | 0.050 | 0.0043 | No | ||
| 28 | GLI2 | 3060632 | 4406 | 0.046 | -0.0031 | No | ||
| 29 | FOXP3 | 940707 1850692 | 4525 | 0.043 | -0.0081 | No | ||
| 30 | PLAGL1 | 3190082 6200193 | 4597 | 0.041 | -0.0107 | No | ||
| 31 | ACVR1 | 6840671 | 4738 | 0.039 | -0.0170 | No | ||
| 32 | FOXO3 | 2510484 4480451 | 4783 | 0.038 | -0.0182 | No | ||
| 33 | THRAP3 | 6180309 | 4835 | 0.037 | -0.0198 | No | ||
| 34 | CITED2 | 5670114 5130088 | 4868 | 0.036 | -0.0203 | No | ||
| 35 | MAP2K3 | 5570193 | 5082 | 0.032 | -0.0308 | No | ||
| 36 | FOXE1 | 4810164 | 5098 | 0.032 | -0.0306 | No | ||
| 37 | UBE2N | 520369 2900047 | 5562 | 0.026 | -0.0548 | No | ||
| 38 | CREB5 | 2320368 | 5656 | 0.024 | -0.0590 | No | ||
| 39 | UTF1 | 5700593 | 5824 | 0.023 | -0.0674 | No | ||
| 40 | IL4 | 6020537 | 6057 | 0.020 | -0.0792 | No | ||
| 41 | ACVRL1 | 3290600 | 6155 | 0.019 | -0.0839 | No | ||
| 42 | GLIS2 | 4150471 4810170 | 6309 | 0.018 | -0.0916 | No | ||
| 43 | TBX5 | 2900132 6370484 | 6316 | 0.018 | -0.0913 | No | ||
| 44 | NUFIP1 | 6350241 | 6390 | 0.017 | -0.0947 | No | ||
| 45 | CDK7 | 2640451 | 7752 | 0.009 | -0.1680 | No | ||
| 46 | NPAS2 | 770670 | 7804 | 0.009 | -0.1704 | No | ||
| 47 | RORB | 6400035 | 7857 | 0.009 | -0.1729 | No | ||
| 48 | MKL2 | 5700204 | 8056 | 0.008 | -0.1834 | No | ||
| 49 | NRIP1 | 3190039 | 8083 | 0.008 | -0.1845 | No | ||
| 50 | FOXF2 | 1240091 | 8114 | 0.008 | -0.1859 | No | ||
| 51 | GATA4 | 1410215 2690609 | 8594 | 0.006 | -0.2116 | No | ||
| 52 | ABCA1 | 6290156 | 8669 | 0.006 | -0.2154 | No | ||
| 53 | HIF1A | 5670605 | 8842 | 0.005 | -0.2245 | No | ||
| 54 | ESRRG | 4010181 | 9935 | 0.002 | -0.2835 | No | ||
| 55 | EHF | 4560088 | 9976 | 0.002 | -0.2856 | No | ||
| 56 | IRF4 | 610133 | 10272 | 0.001 | -0.3015 | No | ||
| 57 | SMARCA1 | 4230315 | 10479 | 0.001 | -0.3126 | No | ||
| 58 | EREG | 50519 4920129 | 10683 | 0.000 | -0.3235 | No | ||
| 59 | ARNT2 | 1740594 | 11133 | -0.001 | -0.3478 | No | ||
| 60 | BMP6 | 2100128 | 11286 | -0.002 | -0.3559 | No | ||
| 61 | NCOA6 | 1780333 6450110 | 11459 | -0.002 | -0.3652 | No | ||
| 62 | MYST4 | 1400563 2570687 3360458 6840402 | 11916 | -0.004 | -0.3897 | No | ||
| 63 | GLI1 | 4560176 | 11972 | -0.004 | -0.3925 | No | ||
| 64 | FOXD3 | 6550156 | 12005 | -0.004 | -0.3941 | No | ||
| 65 | FOXH1 | 2060180 | 12354 | -0.005 | -0.4128 | No | ||
| 66 | RGMB | 670504 4570450 | 12529 | -0.006 | -0.4220 | No | ||
| 67 | YWHAH | 1660133 2810053 | 12889 | -0.008 | -0.4412 | No | ||
| 68 | PAX8 | 5050301 5290164 | 13741 | -0.014 | -0.4867 | No | ||
| 69 | PPARGC1A | 4670040 | 13963 | -0.016 | -0.4982 | No | ||
| 70 | BPTF | 940541 4540152 | 14159 | -0.019 | -0.5081 | No | ||
| 71 | CLOCK | 1410070 2940292 3390075 | 14298 | -0.021 | -0.5149 | No | ||
| 72 | NFATC2 | 70450 540097 | 14309 | -0.021 | -0.5148 | No | ||
| 73 | RXRA | 3800471 | 14505 | -0.024 | -0.5245 | No | ||
| 74 | TCF3 | 5080022 | 14733 | -0.029 | -0.5359 | No | ||
| 75 | NOTCH4 | 2450040 6370707 | 14957 | -0.035 | -0.5468 | No | ||
| 76 | RAD51 | 6110450 6980280 | 15032 | -0.039 | -0.5496 | No | ||
| 77 | GLIS1 | 840471 | 15182 | -0.045 | -0.5562 | No | ||
| 78 | UBB | 4810138 | 15183 | -0.045 | -0.5548 | No | ||
| 79 | ELF4 | 4850193 | 15444 | -0.057 | -0.5670 | No | ||
| 80 | HIVEP3 | 3990551 | 15632 | -0.069 | -0.5749 | No | ||
| 81 | NARG1 | 5910563 6350095 | 15804 | -0.081 | -0.5816 | No | ||
| 82 | BCL10 | 2360397 | 15829 | -0.083 | -0.5802 | No | ||
| 83 | TRERF1 | 6370017 6940138 | 15841 | -0.084 | -0.5781 | No | ||
| 84 | ERCC3 | 6900008 | 15992 | -0.095 | -0.5832 | No | ||
| 85 | NSD1 | 4560519 4780672 | 16074 | -0.102 | -0.5843 | No | ||
| 86 | YAF2 | 940053 | 16441 | -0.141 | -0.5996 | No | ||
| 87 | PPARGC1B | 3290114 | 16512 | -0.152 | -0.5985 | No | ||
| 88 | MYO6 | 2190332 | 16625 | -0.169 | -0.5992 | No | ||
| 89 | BCL3 | 3990440 | 16752 | -0.187 | -0.6000 | Yes | ||
| 90 | MED6 | 6840500 | 16755 | -0.187 | -0.5941 | Yes | ||
| 91 | SP1 | 6590017 | 16838 | -0.205 | -0.5920 | Yes | ||
| 92 | ARNTL | 3170463 | 16863 | -0.209 | -0.5867 | Yes | ||
| 93 | PHF5A | 2690519 | 16977 | -0.228 | -0.5855 | Yes | ||
| 94 | TNFRSF1A | 1090390 6520735 | 17180 | -0.268 | -0.5879 | Yes | ||
| 95 | ILF2 | 2900253 | 17184 | -0.270 | -0.5794 | Yes | ||
| 96 | SMAD2 | 4200592 | 17271 | -0.295 | -0.5746 | Yes | ||
| 97 | MNAT1 | 730292 2350364 | 17289 | -0.300 | -0.5660 | Yes | ||
| 98 | CREBBP | 5690035 7040050 | 17381 | -0.327 | -0.5605 | Yes | ||
| 99 | TCF4 | 520021 | 17388 | -0.328 | -0.5504 | Yes | ||
| 100 | HNRPAB | 540504 | 17390 | -0.328 | -0.5399 | Yes | ||
| 101 | MAML1 | 2760008 | 17520 | -0.360 | -0.5354 | Yes | ||
| 102 | GTF2H1 | 2570746 2650148 | 17587 | -0.381 | -0.5268 | Yes | ||
| 103 | MED12 | 4920095 | 17715 | -0.430 | -0.5199 | Yes | ||
| 104 | RSF1 | 1580097 | 17746 | -0.443 | -0.5074 | Yes | ||
| 105 | RUNX1 | 3840711 | 18014 | -0.563 | -0.5039 | Yes | ||
| 106 | ARHGEF11 | 1780288 | 18092 | -0.610 | -0.4885 | Yes | ||
| 107 | CCNH | 3190347 | 18256 | -0.727 | -0.4742 | Yes | ||
| 108 | ATF7IP | 2690176 6770021 | 18287 | -0.759 | -0.4516 | Yes | ||
| 109 | SMARCC1 | 5080019 7100047 | 18369 | -0.848 | -0.4289 | Yes | ||
| 110 | XRCC6 | 2850450 3870528 | 18427 | -0.927 | -0.4024 | Yes | ||
| 111 | CTCF | 5340017 | 18434 | -0.936 | -0.3728 | Yes | ||
| 112 | ELF1 | 4780450 | 18546 | -1.372 | -0.3350 | Yes | ||
| 113 | ILF3 | 940722 3190647 6520110 | 18599 | -2.087 | -0.2712 | Yes | ||
| 114 | ATF4 | 730441 1740195 | 18600 | -2.197 | -0.2011 | Yes | ||
| 115 | ARID1A | 2630022 1690551 4810110 | 18616 | -6.327 | 0.0000 | Yes |

