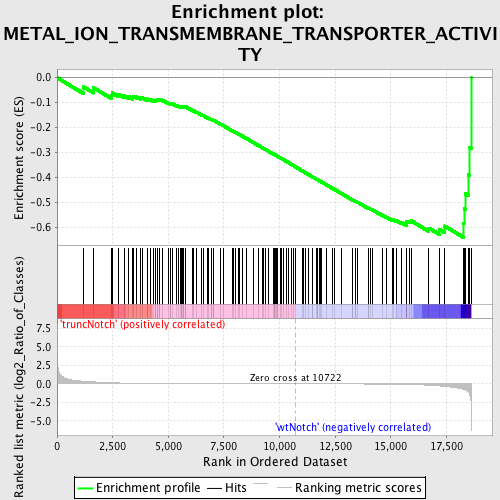
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | METAL_ION_TRANSMEMBRANE_TRANSPORTER_ACTIVITY |
| Enrichment Score (ES) | -0.64170957 |
| Normalized Enrichment Score (NES) | -1.6700209 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.116817884 |
| FWER p-Value | 0.493 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SLC11A2 | 3140603 | 1191 | 0.356 | -0.0370 | No | ||
| 2 | FXN | 4070500 | 1649 | 0.268 | -0.0411 | No | ||
| 3 | ATP2A1 | 110309 | 2443 | 0.159 | -0.0718 | No | ||
| 4 | KCNMB4 | 3290347 | 2469 | 0.156 | -0.0612 | No | ||
| 5 | KCNQ1 | 1580050 6860441 | 2753 | 0.128 | -0.0666 | No | ||
| 6 | ITPR3 | 4010632 | 3022 | 0.105 | -0.0730 | No | ||
| 7 | P2RX4 | 2060497 6650162 | 3208 | 0.094 | -0.0758 | No | ||
| 8 | KCNE2 | 1780577 | 3400 | 0.082 | -0.0799 | No | ||
| 9 | KCNC3 | 3440026 4670288 | 3418 | 0.081 | -0.0746 | No | ||
| 10 | ACCN4 | 6380551 | 3573 | 0.073 | -0.0772 | No | ||
| 11 | KCNJ14 | 870441 4060113 6110372 | 3741 | 0.067 | -0.0811 | No | ||
| 12 | KCNJ6 | 580465 5570246 | 3821 | 0.064 | -0.0805 | No | ||
| 13 | KCNS1 | 2570195 | 4043 | 0.056 | -0.0881 | No | ||
| 14 | CACNA1F | 2360605 3390450 | 4075 | 0.055 | -0.0856 | No | ||
| 15 | CHRNE | 3190170 | 4186 | 0.052 | -0.0876 | No | ||
| 16 | TRPM2 | 460593 | 4337 | 0.047 | -0.0920 | No | ||
| 17 | TRPM1 | 50746 2060301 5130100 5720129 | 4433 | 0.045 | -0.0937 | No | ||
| 18 | SCNN1B | 5390377 | 4439 | 0.045 | -0.0905 | No | ||
| 19 | SCN1B | 1240022 2120048 | 4499 | 0.044 | -0.0903 | No | ||
| 20 | HTR3A | 3310180 | 4523 | 0.043 | -0.0882 | No | ||
| 21 | CACNG1 | 3060711 | 4601 | 0.041 | -0.0892 | No | ||
| 22 | KCNA3 | 510059 | 4621 | 0.041 | -0.0871 | No | ||
| 23 | CHRNA7 | 2970446 | 4749 | 0.038 | -0.0910 | No | ||
| 24 | KCNE1 | 1230520 1690372 4120050 | 5005 | 0.034 | -0.1022 | No | ||
| 25 | ABCC8 | 2810671 3120671 | 5084 | 0.032 | -0.1039 | No | ||
| 26 | KCNH3 | 3130100 | 5103 | 0.032 | -0.1024 | No | ||
| 27 | TRPC5 | 3390270 | 5205 | 0.031 | -0.1056 | No | ||
| 28 | KCNQ3 | 1240037 | 5385 | 0.028 | -0.1131 | No | ||
| 29 | CACNA1S | 4730465 | 5471 | 0.027 | -0.1156 | No | ||
| 30 | SLC8A1 | 3440717 | 5553 | 0.026 | -0.1180 | No | ||
| 31 | KCNH1 | 4070619 6660181 | 5596 | 0.025 | -0.1183 | No | ||
| 32 | CACNA2D1 | 5550593 | 5603 | 0.025 | -0.1167 | No | ||
| 33 | CHRNA3 | 6760100 | 5638 | 0.025 | -0.1167 | No | ||
| 34 | TRPV6 | 4010717 | 5658 | 0.024 | -0.1158 | No | ||
| 35 | KCNA6 | 450291 1400086 1660291 2470576 | 5660 | 0.024 | -0.1140 | No | ||
| 36 | CHRNB3 | 430441 870725 5720204 | 5762 | 0.023 | -0.1176 | No | ||
| 37 | CHRNB4 | 6370110 | 5789 | 0.023 | -0.1173 | No | ||
| 38 | KCNJ10 | 2260577 | 6099 | 0.020 | -0.1324 | No | ||
| 39 | SLC40A1 | 2370286 | 6149 | 0.019 | -0.1336 | No | ||
| 40 | CHRNA2 | 5050315 | 6271 | 0.018 | -0.1387 | No | ||
| 41 | CACNA1G | 3060161 4810706 5550722 | 6473 | 0.017 | -0.1483 | No | ||
| 42 | CUL5 | 450142 | 6562 | 0.016 | -0.1518 | No | ||
| 43 | CACNA1B | 1740010 5360593 | 6760 | 0.015 | -0.1614 | No | ||
| 44 | SCNN1G | 520600 | 6802 | 0.014 | -0.1625 | No | ||
| 45 | KCNK7 | 4230576 | 6932 | 0.014 | -0.1684 | No | ||
| 46 | TRPC1 | 3120189 4210537 | 6959 | 0.013 | -0.1688 | No | ||
| 47 | ABCB11 | 7040170 | 7023 | 0.013 | -0.1712 | No | ||
| 48 | KCNK4 | 3170411 7040064 | 7028 | 0.013 | -0.1704 | No | ||
| 49 | KCNJ1 | 610707 870181 | 7343 | 0.011 | -0.1865 | No | ||
| 50 | CLDN16 | 1850286 | 7472 | 0.011 | -0.1926 | No | ||
| 51 | KCNJ15 | 70176 6900750 | 7877 | 0.009 | -0.2137 | No | ||
| 52 | KCNA1 | 2030594 | 7915 | 0.009 | -0.2151 | No | ||
| 53 | P2RX1 | 2940021 3290338 | 8001 | 0.008 | -0.2190 | No | ||
| 54 | CACNG5 | 1410746 2030673 | 8170 | 0.008 | -0.2275 | No | ||
| 55 | CHRNA6 | 5340092 | 8180 | 0.008 | -0.2274 | No | ||
| 56 | MS4A2 | 2940594 | 8314 | 0.007 | -0.2340 | No | ||
| 57 | KCNJ12 | 3870364 | 8510 | 0.007 | -0.2441 | No | ||
| 58 | KCNC1 | 770110 | 8840 | 0.005 | -0.2614 | No | ||
| 59 | TRPC4 | 6040022 | 9051 | 0.005 | -0.2724 | No | ||
| 60 | ACCN1 | 2060139 2190541 | 9213 | 0.004 | -0.2808 | No | ||
| 61 | SCN11A | 2480372 | 9258 | 0.004 | -0.2829 | No | ||
| 62 | ATP2B3 | 2260022 3780059 | 9379 | 0.004 | -0.2891 | No | ||
| 63 | CACNB3 | 2480035 4540184 | 9501 | 0.003 | -0.2953 | No | ||
| 64 | TRPC3 | 840064 | 9706 | 0.003 | -0.3061 | No | ||
| 65 | KCNA4 | 2320706 | 9759 | 0.003 | -0.3087 | No | ||
| 66 | SCN4A | 5340687 | 9787 | 0.003 | -0.3100 | No | ||
| 67 | KCND3 | 670041 | 9800 | 0.003 | -0.3105 | No | ||
| 68 | CCS | 3450524 | 9849 | 0.002 | -0.3129 | No | ||
| 69 | SCN9A | 5270575 | 9890 | 0.002 | -0.3148 | No | ||
| 70 | SLC30A4 | 1850372 | 9891 | 0.002 | -0.3147 | No | ||
| 71 | SLC30A3 | 1850451 | 10057 | 0.002 | -0.3234 | No | ||
| 72 | CHRND | 840403 2260670 | 10101 | 0.002 | -0.3256 | No | ||
| 73 | KCNJ5 | 1570053 | 10177 | 0.001 | -0.3296 | No | ||
| 74 | KCNMB1 | 4760139 | 10316 | 0.001 | -0.3369 | No | ||
| 75 | P2RX3 | 4230397 | 10381 | 0.001 | -0.3403 | No | ||
| 76 | KCNS3 | 4150039 | 10524 | 0.001 | -0.3480 | No | ||
| 77 | KCNA5 | 5270750 | 10637 | 0.000 | -0.3540 | No | ||
| 78 | KCNK1 | 1050068 | 10696 | 0.000 | -0.3571 | No | ||
| 79 | KCNJ4 | 2350315 | 11051 | -0.001 | -0.3762 | No | ||
| 80 | CACNA1E | 840253 | 11094 | -0.001 | -0.3784 | No | ||
| 81 | KCNK3 | 2690372 | 11146 | -0.001 | -0.3811 | No | ||
| 82 | KCNQ2 | 130204 290291 5860132 | 11300 | -0.002 | -0.3892 | No | ||
| 83 | SLC9A7 | 5360301 | 11466 | -0.002 | -0.3980 | No | ||
| 84 | CACNB4 | 2030133 | 11467 | -0.002 | -0.3978 | No | ||
| 85 | CACNB1 | 2940427 3710487 | 11676 | -0.003 | -0.4088 | No | ||
| 86 | CHRNA1 | 1170025 3610364 5220292 | 11687 | -0.003 | -0.4092 | No | ||
| 87 | SLC13A4 | 2940541 | 11720 | -0.003 | -0.4107 | No | ||
| 88 | RYR1 | 2510114 3130408 3710086 | 11793 | -0.003 | -0.4143 | No | ||
| 89 | CACNA1A | 6220164 | 11842 | -0.003 | -0.4167 | No | ||
| 90 | KCNMB2 | 3780128 4760136 | 11862 | -0.003 | -0.4174 | No | ||
| 91 | PKD2 | 4670390 | 12093 | -0.004 | -0.4295 | No | ||
| 92 | KCNIP2 | 60088 1780324 | 12370 | -0.005 | -0.4441 | No | ||
| 93 | KCNJ3 | 50184 | 12453 | -0.006 | -0.4480 | No | ||
| 94 | CHRNA4 | 730075 2680091 | 12796 | -0.007 | -0.4660 | No | ||
| 95 | SLC39A2 | 1410338 | 13262 | -0.010 | -0.4904 | No | ||
| 96 | KCNA2 | 5080403 | 13291 | -0.010 | -0.4911 | No | ||
| 97 | RYR2 | 2760671 | 13418 | -0.011 | -0.4970 | No | ||
| 98 | CACNA1D | 1740315 4730731 | 13428 | -0.011 | -0.4967 | No | ||
| 99 | KCNN3 | 6520600 6420138 | 13523 | -0.012 | -0.5008 | No | ||
| 100 | SCN7A | 2510538 | 13981 | -0.016 | -0.5243 | No | ||
| 101 | KCNC4 | 110053 4760600 | 13988 | -0.016 | -0.5234 | No | ||
| 102 | CACNA1C | 1660128 3780300 3870092 6290403 6900301 | 14100 | -0.018 | -0.5280 | No | ||
| 103 | HTR3B | 430438 520301 | 14158 | -0.019 | -0.5297 | No | ||
| 104 | SLC31A2 | 1450577 | 14610 | -0.026 | -0.5520 | No | ||
| 105 | SCN5A | 4070017 | 14818 | -0.032 | -0.5608 | No | ||
| 106 | RYR3 | 1660156 | 15059 | -0.040 | -0.5707 | No | ||
| 107 | KCNK5 | 4590504 6860687 | 15108 | -0.042 | -0.5701 | No | ||
| 108 | ATP2C1 | 2630446 6520253 | 15233 | -0.047 | -0.5732 | No | ||
| 109 | CHRNB2 | 580204 3120739 | 15462 | -0.058 | -0.5811 | No | ||
| 110 | ITPR2 | 5360128 | 15705 | -0.074 | -0.5884 | No | ||
| 111 | NOLA1 | 730093 | 15706 | -0.074 | -0.5828 | No | ||
| 112 | KCNN2 | 990025 3520524 | 15710 | -0.074 | -0.5772 | No | ||
| 113 | KCNN4 | 2680129 | 15821 | -0.083 | -0.5768 | No | ||
| 114 | CACNB2 | 1500095 7330707 | 15912 | -0.089 | -0.5748 | No | ||
| 115 | P2RX7 | 6020048 6380053 6510121 | 16708 | -0.182 | -0.6038 | No | ||
| 116 | KCNH2 | 1170451 | 17179 | -0.268 | -0.6086 | Yes | ||
| 117 | CACNG4 | 6980112 | 17419 | -0.336 | -0.5957 | Yes | ||
| 118 | SLC30A5 | 2630288 2970403 | 18271 | -0.744 | -0.5845 | Yes | ||
| 119 | CACNA1H | 1230279 | 18291 | -0.765 | -0.5266 | Yes | ||
| 120 | ITPR1 | 3450519 | 18349 | -0.823 | -0.4664 | Yes | ||
| 121 | SLC31A1 | 4590161 6940154 | 18492 | -1.090 | -0.3902 | Yes | ||
| 122 | ATP2A2 | 1090075 3990279 | 18554 | -1.475 | -0.2800 | Yes | ||
| 123 | ATP2A3 | 130440 2190451 | 18613 | -3.680 | 0.0002 | Yes |

