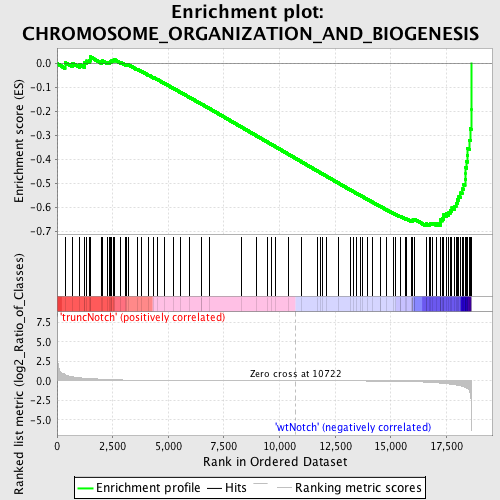
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | CHROMOSOME_ORGANIZATION_AND_BIOGENESIS |
| Enrichment Score (ES) | -0.67725223 |
| Normalized Enrichment Score (NES) | -1.7332028 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.10140965 |
| FWER p-Value | 0.157 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HDAC5 | 2370035 6370139 | 355 | 0.797 | 0.0047 | No | ||
| 2 | ERCC1 | 3800010 4590132 4670397 | 699 | 0.510 | 0.0014 | No | ||
| 3 | CHMP1A | 5550441 | 1022 | 0.394 | -0.0041 | No | ||
| 4 | HMGA1 | 6580408 | 1226 | 0.349 | -0.0046 | No | ||
| 5 | SMARCD1 | 3060193 3850184 6400369 | 1240 | 0.347 | 0.0050 | No | ||
| 6 | SUPT4H1 | 6200056 | 1313 | 0.332 | 0.0111 | No | ||
| 7 | HTATIP | 5360739 | 1453 | 0.303 | 0.0127 | No | ||
| 8 | RBM14 | 5340731 5690315 | 1486 | 0.297 | 0.0198 | No | ||
| 9 | HDAC7A | 2570278 | 1494 | 0.296 | 0.0283 | No | ||
| 10 | SIRT2 | 630494 | 2004 | 0.216 | 0.0073 | No | ||
| 11 | RBBP4 | 6650528 | 2043 | 0.210 | 0.0116 | No | ||
| 12 | MYST3 | 5270500 | 2270 | 0.181 | 0.0048 | No | ||
| 13 | GCN5L2 | 2340609 | 2364 | 0.168 | 0.0048 | No | ||
| 14 | HDAC6 | 6200446 | 2379 | 0.166 | 0.0090 | No | ||
| 15 | HDAC3 | 4060072 | 2427 | 0.161 | 0.0113 | No | ||
| 16 | STAG3 | 3830112 | 2466 | 0.156 | 0.0139 | No | ||
| 17 | AIFM2 | 360537 3780520 4230138 6130082 | 2526 | 0.150 | 0.0152 | No | ||
| 18 | TAF6L | 1850056 | 2588 | 0.144 | 0.0163 | No | ||
| 19 | MYST1 | 2190397 | 2865 | 0.117 | 0.0049 | No | ||
| 20 | MSH2 | 6180273 | 3077 | 0.102 | -0.0035 | No | ||
| 21 | TSSK6 | 2190136 | 3132 | 0.099 | -0.0034 | No | ||
| 22 | TEP1 | 7040735 | 3196 | 0.095 | -0.0040 | No | ||
| 23 | HDAC10 | 2850204 | 3594 | 0.072 | -0.0233 | No | ||
| 24 | CHAF1B | 3940142 | 3771 | 0.066 | -0.0308 | No | ||
| 25 | CHAF1A | 6040647 | 4123 | 0.054 | -0.0482 | No | ||
| 26 | MTA2 | 5360242 5720494 | 4353 | 0.047 | -0.0591 | No | ||
| 27 | JMJD2A | 3610128 4200373 | 4505 | 0.044 | -0.0660 | No | ||
| 28 | TINF2 | 2360358 | 4847 | 0.037 | -0.0833 | No | ||
| 29 | CDCA5 | 5670131 | 5231 | 0.030 | -0.1031 | No | ||
| 30 | UBE2N | 520369 2900047 | 5562 | 0.026 | -0.1201 | No | ||
| 31 | CENPE | 2850022 | 5930 | 0.021 | -0.1393 | No | ||
| 32 | TERT | 6660075 | 6488 | 0.017 | -0.1689 | No | ||
| 33 | NAP1L3 | 6020079 | 6496 | 0.016 | -0.1688 | No | ||
| 34 | SYCP3 | 5420286 | 6839 | 0.014 | -0.1868 | No | ||
| 35 | BNIP3 | 3140270 | 8305 | 0.007 | -0.2657 | No | ||
| 36 | HELLS | 4560086 4810025 | 8978 | 0.005 | -0.3019 | No | ||
| 37 | SET | 6650286 | 9451 | 0.004 | -0.3273 | No | ||
| 38 | PHB | 2260739 | 9622 | 0.003 | -0.3363 | No | ||
| 39 | SUV39H2 | 1450672 6370242 | 9814 | 0.003 | -0.3466 | No | ||
| 40 | DFFB | 1770092 | 10383 | 0.001 | -0.3772 | No | ||
| 41 | MAP3K12 | 2470373 6420162 | 10996 | -0.001 | -0.4103 | No | ||
| 42 | POT1 | 5340184 | 11709 | -0.003 | -0.4486 | No | ||
| 43 | PAM | 5290528 | 11858 | -0.003 | -0.4565 | No | ||
| 44 | MYST4 | 1400563 2570687 3360458 6840402 | 11916 | -0.004 | -0.4595 | No | ||
| 45 | NAP1L2 | 130047 | 12094 | -0.004 | -0.4689 | No | ||
| 46 | HMGB1 | 2120670 2350044 | 12631 | -0.006 | -0.4977 | No | ||
| 47 | TNP1 | 3120048 | 13190 | -0.009 | -0.5275 | No | ||
| 48 | NBN | 730538 2470619 4780594 | 13305 | -0.010 | -0.5334 | No | ||
| 49 | CDC23 | 3190593 | 13446 | -0.011 | -0.5406 | No | ||
| 50 | HMGA2 | 2940121 3390647 5130279 6400136 | 13652 | -0.013 | -0.5513 | No | ||
| 51 | SYCP1 | 2570010 | 13740 | -0.014 | -0.5556 | No | ||
| 52 | PPARGC1A | 4670040 | 13963 | -0.016 | -0.5671 | No | ||
| 53 | BPTF | 940541 4540152 | 14159 | -0.019 | -0.5771 | No | ||
| 54 | HDAC8 | 3440504 | 14532 | -0.025 | -0.5964 | No | ||
| 55 | NPM2 | 6040139 | 14553 | -0.025 | -0.5967 | No | ||
| 56 | DDX11 | 6590671 | 14823 | -0.032 | -0.6103 | No | ||
| 57 | SIRT5 | 110075 2640025 | 15122 | -0.042 | -0.6251 | No | ||
| 58 | HDAC11 | 2510242 | 15228 | -0.046 | -0.6294 | No | ||
| 59 | RPS6KA5 | 2120563 7040546 | 15421 | -0.055 | -0.6381 | No | ||
| 60 | PTGES3 | 2190440 2680524 | 15425 | -0.056 | -0.6366 | No | ||
| 61 | SMARCA5 | 6620050 | 15678 | -0.072 | -0.6481 | No | ||
| 62 | TERF2IP | 580010 870364 2190358 | 15718 | -0.075 | -0.6479 | No | ||
| 63 | SMC4 | 5910240 | 15937 | -0.091 | -0.6570 | No | ||
| 64 | SMG6 | 5820243 | 15986 | -0.095 | -0.6567 | No | ||
| 65 | CARM1 | 7040292 | 15993 | -0.096 | -0.6542 | No | ||
| 66 | MSH3 | 6770575 | 15995 | -0.096 | -0.6514 | No | ||
| 67 | RAD50 | 4050184 6770746 | 16046 | -0.100 | -0.6511 | No | ||
| 68 | NSD1 | 4560519 4780672 | 16074 | -0.102 | -0.6495 | No | ||
| 69 | ASF1A | 7050020 | 16589 | -0.163 | -0.6724 | Yes | ||
| 70 | NAP1L1 | 2510593 5900215 5090470 450079 4920280 | 16594 | -0.164 | -0.6677 | Yes | ||
| 71 | SMARCE1 | 4920465 4390398 | 16743 | -0.186 | -0.6701 | Yes | ||
| 72 | EZH2 | 6130605 6380524 | 16764 | -0.189 | -0.6655 | Yes | ||
| 73 | SIRT1 | 1190731 | 16889 | -0.212 | -0.6659 | Yes | ||
| 74 | CENPH | 4280097 | 17065 | -0.245 | -0.6680 | Yes | ||
| 75 | HDAC1 | 2850670 | 17220 | -0.280 | -0.6679 | Yes | ||
| 76 | NDC80 | 4120465 | 17235 | -0.283 | -0.6601 | Yes | ||
| 77 | ZWINT | 6940670 | 17240 | -0.284 | -0.6518 | Yes | ||
| 78 | TLK2 | 1170161 | 17328 | -0.312 | -0.6472 | Yes | ||
| 79 | TERF2 | 3840044 | 17372 | -0.324 | -0.6398 | Yes | ||
| 80 | CREBBP | 5690035 7040050 | 17381 | -0.327 | -0.6305 | Yes | ||
| 81 | NAP1L4 | 1240504 | 17495 | -0.354 | -0.6260 | Yes | ||
| 82 | ACIN1 | 3390278 6350377 6620142 | 17608 | -0.389 | -0.6204 | Yes | ||
| 83 | POLS | 5890438 | 17665 | -0.415 | -0.6110 | Yes | ||
| 84 | RSF1 | 1580097 | 17746 | -0.443 | -0.6020 | Yes | ||
| 85 | SUPT16H | 1240333 | 17875 | -0.491 | -0.5942 | Yes | ||
| 86 | MRE11A | 3850601 4670332 | 17933 | -0.523 | -0.5816 | Yes | ||
| 87 | ERCC4 | 1570619 5720180 | 17986 | -0.547 | -0.5680 | Yes | ||
| 88 | RFC1 | 1190673 | 18057 | -0.589 | -0.5541 | Yes | ||
| 89 | NCAPH | 6220435 | 18140 | -0.637 | -0.5395 | Yes | ||
| 90 | NASP | 2260139 2940369 5130707 | 18205 | -0.676 | -0.5227 | Yes | ||
| 91 | TOP2A | 360717 | 18276 | -0.748 | -0.5041 | Yes | ||
| 92 | ZW10 | 2900735 3520687 | 18335 | -0.804 | -0.4831 | Yes | ||
| 93 | EHMT1 | 5290433 5910086 6620112 6980014 | 18364 | -0.844 | -0.4594 | Yes | ||
| 94 | SMARCC1 | 5080019 7100047 | 18369 | -0.848 | -0.4342 | Yes | ||
| 95 | TLK1 | 7100605 | 18412 | -0.907 | -0.4092 | Yes | ||
| 96 | SATB1 | 5670154 | 18438 | -0.951 | -0.3821 | Yes | ||
| 97 | HDAC2 | 4050433 | 18455 | -0.999 | -0.3530 | Yes | ||
| 98 | SMC1A | 3060600 5700148 5890113 6370154 | 18529 | -1.261 | -0.3192 | Yes | ||
| 99 | ACTL6A | 5220543 6290541 2450093 4590332 5270722 | 18570 | -1.652 | -0.2719 | Yes | ||
| 100 | NUSAP1 | 940048 3120435 | 18611 | -2.829 | -0.1893 | Yes | ||
| 101 | ARID1A | 2630022 1690551 4810110 | 18616 | -6.327 | 0.0000 | Yes |

