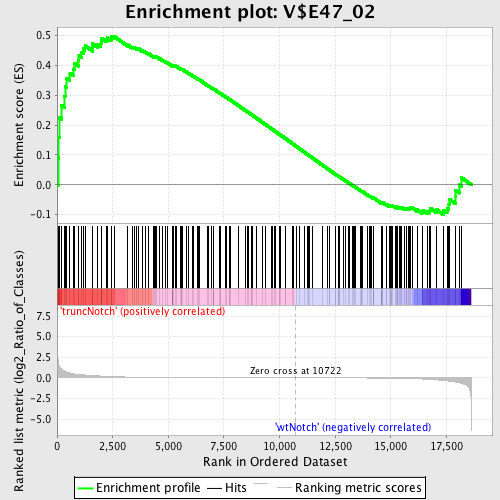
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | truncNotch |
| GeneSet | V$E47_02 |
| Enrichment Score (ES) | 0.49818626 |
| Normalized Enrichment Score (NES) | 1.2830966 |
| Nominal p-value | 0.03776683 |
| FDR q-value | 0.9059407 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | EPHB6 | 5860128 | 44 | 2.078 | 0.0901 | Yes | ||
| 2 | NCDN | 50040 1980739 3520603 | 77 | 1.599 | 0.1595 | Yes | ||
| 3 | HES6 | 540411 6550504 | 96 | 1.504 | 0.2255 | Yes | ||
| 4 | PADI4 | 1740075 4810441 | 204 | 1.039 | 0.2660 | Yes | ||
| 5 | TPI1 | 1500215 2100154 | 337 | 0.825 | 0.2955 | Yes | ||
| 6 | AIG1 | 2450091 | 357 | 0.795 | 0.3299 | Yes | ||
| 7 | POU2AF1 | 5690242 | 427 | 0.694 | 0.3570 | Yes | ||
| 8 | DGKA | 5720152 5890328 | 578 | 0.577 | 0.3746 | Yes | ||
| 9 | GNA12 | 1230301 | 737 | 0.495 | 0.3880 | Yes | ||
| 10 | TNFSF13 | 2650170 | 776 | 0.479 | 0.4073 | Yes | ||
| 11 | RAB33A | 6590195 | 950 | 0.417 | 0.4165 | Yes | ||
| 12 | TRIB2 | 4120605 | 982 | 0.408 | 0.4330 | Yes | ||
| 13 | IRF1 | 2340152 3450592 6290121 6980577 | 1095 | 0.374 | 0.4435 | Yes | ||
| 14 | SASH1 | 4070270 | 1172 | 0.358 | 0.4554 | Yes | ||
| 15 | MGAT1 | 2450377 | 1253 | 0.344 | 0.4663 | Yes | ||
| 16 | CUEDC1 | 1660463 | 1591 | 0.278 | 0.4604 | Yes | ||
| 17 | PARD6A | 4120170 | 1600 | 0.276 | 0.4723 | Yes | ||
| 18 | ARID5A | 2450133 | 1827 | 0.242 | 0.4708 | Yes | ||
| 19 | ARMCX2 | 2320167 | 1972 | 0.219 | 0.4728 | Yes | ||
| 20 | DULLARD | 130180 | 1980 | 0.219 | 0.4821 | Yes | ||
| 21 | LMO2 | 5360450 | 2009 | 0.215 | 0.4902 | Yes | ||
| 22 | STC2 | 4920601 | 2223 | 0.187 | 0.4869 | Yes | ||
| 23 | MYO18A | 1230333 | 2266 | 0.181 | 0.4927 | Yes | ||
| 24 | HDAC3 | 4060072 | 2427 | 0.161 | 0.4912 | Yes | ||
| 25 | BCL9 | 7100112 | 2431 | 0.161 | 0.4982 | Yes | ||
| 26 | CRSP7 | 4230408 | 2561 | 0.147 | 0.4977 | No | ||
| 27 | PRELP | 5900390 | 3174 | 0.096 | 0.4688 | No | ||
| 28 | NUMBL | 6130360 6200333 | 3376 | 0.083 | 0.4616 | No | ||
| 29 | TPM2 | 520735 3870390 | 3478 | 0.078 | 0.4596 | No | ||
| 30 | ACCN4 | 6380551 | 3573 | 0.073 | 0.4578 | No | ||
| 31 | ALX3 | 50497 | 3654 | 0.070 | 0.4566 | No | ||
| 32 | FGF13 | 630575 1570440 5360121 | 3845 | 0.063 | 0.4490 | No | ||
| 33 | CPB1 | 290600 | 3975 | 0.058 | 0.4447 | No | ||
| 34 | MAPK12 | 450022 1340717 7050484 | 4113 | 0.054 | 0.4396 | No | ||
| 35 | PLEC1 | 6520292 50500 380075 1570088 2510692 2630497 2680017 2900746 4560497 4850376 6290398 7040471 | 4349 | 0.047 | 0.4290 | No | ||
| 36 | BRUNOL4 | 4490452 5080451 | 4388 | 0.046 | 0.4290 | No | ||
| 37 | A2BP1 | 2370390 4590593 5550014 | 4412 | 0.046 | 0.4298 | No | ||
| 38 | WDR6 | 7050154 | 4470 | 0.045 | 0.4286 | No | ||
| 39 | SYTL2 | 580097 5390576 6770603 | 4615 | 0.041 | 0.4227 | No | ||
| 40 | DDAH2 | 4480551 | 4757 | 0.038 | 0.4167 | No | ||
| 41 | CASKIN2 | 2030113 | 4884 | 0.036 | 0.4115 | No | ||
| 42 | CEL | 2640278 | 4963 | 0.034 | 0.4088 | No | ||
| 43 | SNAP25 | 360520 | 5174 | 0.031 | 0.3988 | No | ||
| 44 | MLLT6 | 3870168 | 5190 | 0.031 | 0.3993 | No | ||
| 45 | POU3F2 | 6620484 | 5191 | 0.031 | 0.4007 | No | ||
| 46 | MEF2D | 5690576 | 5207 | 0.031 | 0.4013 | No | ||
| 47 | MYOCD | 2650064 3440403 6220121 | 5250 | 0.030 | 0.4003 | No | ||
| 48 | NOL4 | 5050446 5360519 | 5301 | 0.029 | 0.3989 | No | ||
| 49 | CUGBP1 | 450292 510022 7050176 7050215 | 5365 | 0.028 | 0.3967 | No | ||
| 50 | PCDHA6 | 780333 2370280 | 5532 | 0.026 | 0.3889 | No | ||
| 51 | PCSK4 | 130687 670711 3170075 6400411 | 5576 | 0.026 | 0.3877 | No | ||
| 52 | HSD3B7 | 1410129 | 5598 | 0.025 | 0.3877 | No | ||
| 53 | BZW2 | 940079 | 5640 | 0.025 | 0.3866 | No | ||
| 54 | UBXD3 | 5050193 | 5818 | 0.023 | 0.3780 | No | ||
| 55 | ELAVL2 | 360181 | 5914 | 0.022 | 0.3738 | No | ||
| 56 | RIMS2 | 670725 | 6075 | 0.020 | 0.3660 | No | ||
| 57 | AP4S1 | 4730292 | 6135 | 0.020 | 0.3637 | No | ||
| 58 | DMD | 1740041 3990332 | 6288 | 0.018 | 0.3563 | No | ||
| 59 | CALB1 | 460070 | 6374 | 0.018 | 0.3524 | No | ||
| 60 | PLEKHB1 | 3120039 4050270 540546 870463 | 6393 | 0.017 | 0.3522 | No | ||
| 61 | ERBB3 | 5890372 | 6738 | 0.015 | 0.3342 | No | ||
| 62 | CNTN6 | 2630711 | 6798 | 0.014 | 0.3317 | No | ||
| 63 | KLHL1 | 6350021 | 6961 | 0.013 | 0.3235 | No | ||
| 64 | OFCC1 | 2060392 | 7016 | 0.013 | 0.3211 | No | ||
| 65 | GJA4 | 6380452 | 7037 | 0.013 | 0.3206 | No | ||
| 66 | DRD3 | 4780402 | 7048 | 0.013 | 0.3207 | No | ||
| 67 | ABR | 610079 1170609 3610195 5670050 | 7306 | 0.012 | 0.3072 | No | ||
| 68 | MEF2C | 670025 780338 | 7333 | 0.011 | 0.3063 | No | ||
| 69 | ANXA8 | 4780022 | 7357 | 0.011 | 0.3056 | No | ||
| 70 | MESDC1 | 2030019 | 7568 | 0.010 | 0.2947 | No | ||
| 71 | HSPB3 | 460129 | 7622 | 0.010 | 0.2923 | No | ||
| 72 | BNC2 | 4810603 | 7732 | 0.010 | 0.2868 | No | ||
| 73 | FOXA1 | 3710609 | 7746 | 0.009 | 0.2865 | No | ||
| 74 | NPAS2 | 770670 | 7804 | 0.009 | 0.2838 | No | ||
| 75 | ATOH7 | 5550121 | 8156 | 0.008 | 0.2651 | No | ||
| 76 | TTN | 2320161 4670056 6550026 | 8482 | 0.007 | 0.2478 | No | ||
| 77 | KLK1 | 6040369 | 8561 | 0.006 | 0.2439 | No | ||
| 78 | HDAC9 | 1990010 2260133 | 8603 | 0.006 | 0.2419 | No | ||
| 79 | GABRE | 4780020 | 8736 | 0.006 | 0.2350 | No | ||
| 80 | BTBD11 | 6650154 | 8739 | 0.006 | 0.2352 | No | ||
| 81 | SNCAIP | 4010551 | 8769 | 0.006 | 0.2338 | No | ||
| 82 | PCDHA10 | 3830725 | 8970 | 0.005 | 0.2232 | No | ||
| 83 | ACCN1 | 2060139 2190541 | 9213 | 0.004 | 0.2103 | No | ||
| 84 | JPH3 | 780397 | 9344 | 0.004 | 0.2034 | No | ||
| 85 | AKT2 | 3850541 3870519 | 9643 | 0.003 | 0.1874 | No | ||
| 86 | ARL4A | 6290091 | 9685 | 0.003 | 0.1853 | No | ||
| 87 | KCNA4 | 2320706 | 9759 | 0.003 | 0.1815 | No | ||
| 88 | LRRN1 | 3290154 | 9805 | 0.003 | 0.1791 | No | ||
| 89 | PGM3 | 2570465 | 9977 | 0.002 | 0.1700 | No | ||
| 90 | BTBD3 | 1980041 2510577 3190053 4200746 | 9988 | 0.002 | 0.1695 | No | ||
| 91 | DDAH1 | 6400750 | 10019 | 0.002 | 0.1680 | No | ||
| 92 | FOXI1 | 3710095 6620195 | 10244 | 0.001 | 0.1559 | No | ||
| 93 | NGFR | 3140102 | 10282 | 0.001 | 0.1539 | No | ||
| 94 | CHRM1 | 4280619 | 10570 | 0.000 | 0.1384 | No | ||
| 95 | EDG1 | 4200619 | 10622 | 0.000 | 0.1356 | No | ||
| 96 | SERPINI2 | 1570427 | 10742 | -0.000 | 0.1292 | No | ||
| 97 | NOV | 2850746 | 10883 | -0.000 | 0.1216 | No | ||
| 98 | ASB5 | 2760575 | 11107 | -0.001 | 0.1096 | No | ||
| 99 | MMP16 | 2680139 | 11236 | -0.001 | 0.1027 | No | ||
| 100 | FGF11 | 840292 | 11319 | -0.002 | 0.0983 | No | ||
| 101 | LRRN3 | 1450133 | 11335 | -0.002 | 0.0976 | No | ||
| 102 | SLC9A7 | 5360301 | 11466 | -0.002 | 0.0906 | No | ||
| 103 | PRKCQ | 2260170 3870193 | 11911 | -0.004 | 0.0667 | No | ||
| 104 | DOC2B | 2190021 2850142 | 12137 | -0.004 | 0.0547 | No | ||
| 105 | MYC | 380541 4670170 | 12225 | -0.005 | 0.0502 | No | ||
| 106 | BMX | 50136 2940242 4540059 | 12264 | -0.005 | 0.0484 | No | ||
| 107 | IRX4 | 1980102 | 12506 | -0.006 | 0.0356 | No | ||
| 108 | GFAP | 2060092 | 12518 | -0.006 | 0.0352 | No | ||
| 109 | GRIA1 | 1340152 3780750 4920440 | 12645 | -0.007 | 0.0287 | No | ||
| 110 | TCF2 | 870338 5050632 | 12666 | -0.007 | 0.0279 | No | ||
| 111 | PDE4B | 4480121 | 12671 | -0.007 | 0.0280 | No | ||
| 112 | PURA | 3870156 | 12700 | -0.007 | 0.0268 | No | ||
| 113 | NPTX2 | 70021 | 12852 | -0.008 | 0.0189 | No | ||
| 114 | DSCAML1 | 1850403 5550079 | 12963 | -0.008 | 0.0133 | No | ||
| 115 | PFTK1 | 1780181 | 13095 | -0.009 | 0.0066 | No | ||
| 116 | SDPR | 3360292 | 13162 | -0.009 | 0.0034 | No | ||
| 117 | KLHL13 | 6590731 | 13281 | -0.010 | -0.0025 | No | ||
| 118 | FGD4 | 520168 870411 2640253 6550338 6650364 | 13313 | -0.010 | -0.0037 | No | ||
| 119 | DSCAM | 1780050 2450731 2810438 | 13347 | -0.011 | -0.0050 | No | ||
| 120 | FUT8 | 1340068 2340056 | 13420 | -0.011 | -0.0084 | No | ||
| 121 | RRM2B | 1690025 1690465 4010400 | 13649 | -0.013 | -0.0202 | No | ||
| 122 | TEAD2 | 6450079 | 13687 | -0.013 | -0.0217 | No | ||
| 123 | PCDHA1 | 940026 540484 | 13688 | -0.013 | -0.0211 | No | ||
| 124 | DUSP9 | 630605 | 13735 | -0.014 | -0.0230 | No | ||
| 125 | PCM1 | 1940070 3940707 | 13936 | -0.016 | -0.0331 | No | ||
| 126 | HAPLN2 | 2900647 | 14037 | -0.017 | -0.0378 | No | ||
| 127 | RHOBTB1 | 1410010 | 14088 | -0.018 | -0.0397 | No | ||
| 128 | TAL1 | 7040239 | 14131 | -0.018 | -0.0412 | No | ||
| 129 | GATM | 380671 | 14206 | -0.019 | -0.0443 | No | ||
| 130 | JMJD1C | 940575 2120025 | 14213 | -0.019 | -0.0438 | No | ||
| 131 | WNT10B | 510050 | 14230 | -0.020 | -0.0438 | No | ||
| 132 | CDH7 | 6620722 | 14579 | -0.026 | -0.0615 | No | ||
| 133 | CPNE1 | 3440131 | 14581 | -0.026 | -0.0604 | No | ||
| 134 | HOXA7 | 5910152 | 14596 | -0.026 | -0.0600 | No | ||
| 135 | SOX12 | 1780037 | 14611 | -0.026 | -0.0596 | No | ||
| 136 | SLITRK3 | 5910041 | 14641 | -0.027 | -0.0600 | No | ||
| 137 | SEMA4G | 5890026 | 14791 | -0.031 | -0.0667 | No | ||
| 138 | SCN5A | 4070017 | 14818 | -0.032 | -0.0667 | No | ||
| 139 | ASXL2 | 130670 | 14941 | -0.035 | -0.0717 | No | ||
| 140 | PODXL | 130717 | 14981 | -0.037 | -0.0722 | No | ||
| 141 | MKL1 | 6840161 | 14993 | -0.037 | -0.0712 | No | ||
| 142 | TRIM23 | 580168 6510672 | 15009 | -0.038 | -0.0703 | No | ||
| 143 | ADAM12 | 3390132 4070347 | 15020 | -0.038 | -0.0691 | No | ||
| 144 | BAZ2A | 730184 | 15091 | -0.041 | -0.0711 | No | ||
| 145 | ERG | 50154 1770739 | 15200 | -0.045 | -0.0750 | No | ||
| 146 | SHOX2 | 3190438 6450059 | 15270 | -0.048 | -0.0765 | No | ||
| 147 | UNC5B | 2370279 | 15297 | -0.049 | -0.0758 | No | ||
| 148 | RWDD2 | 1660079 | 15310 | -0.050 | -0.0742 | No | ||
| 149 | USP54 | 3060079 | 15377 | -0.053 | -0.0754 | No | ||
| 150 | GALR3 | 4850113 | 15434 | -0.056 | -0.0760 | No | ||
| 151 | THRA | 6770341 110164 1990600 | 15479 | -0.059 | -0.0757 | No | ||
| 152 | HIVEP3 | 3990551 | 15632 | -0.069 | -0.0809 | No | ||
| 153 | STK3 | 7100427 | 15684 | -0.072 | -0.0805 | No | ||
| 154 | KCNN2 | 990025 3520524 | 15710 | -0.074 | -0.0785 | No | ||
| 155 | STRN3 | 1450093 6200685 | 15810 | -0.081 | -0.0802 | No | ||
| 156 | TRERF1 | 6370017 6940138 | 15841 | -0.084 | -0.0781 | No | ||
| 157 | AMPH | 2260338 3140576 | 15861 | -0.085 | -0.0754 | No | ||
| 158 | CHRNB1 | 1190671 | 15952 | -0.092 | -0.0762 | No | ||
| 159 | ART1 | 450288 4570292 | 16179 | -0.112 | -0.0834 | No | ||
| 160 | LYL1 | 2900242 | 16403 | -0.136 | -0.0894 | No | ||
| 161 | PSMD3 | 1400647 | 16442 | -0.142 | -0.0852 | No | ||
| 162 | HMGN2 | 3140091 | 16644 | -0.172 | -0.0884 | No | ||
| 163 | BMF | 610102 | 16757 | -0.188 | -0.0862 | No | ||
| 164 | SLC7A11 | 2850138 | 16767 | -0.190 | -0.0782 | No | ||
| 165 | NR2F6 | 1340239 5670239 60338 4280088 | 17046 | -0.241 | -0.0826 | No | ||
| 166 | BCL2L1 | 1580452 4200152 5420484 | 17359 | -0.321 | -0.0852 | No | ||
| 167 | NDUFS4 | 510142 1240148 4760253 | 17527 | -0.363 | -0.0781 | No | ||
| 168 | IDE | 670746 | 17610 | -0.391 | -0.0651 | No | ||
| 169 | TSGA14 | 6660465 | 17647 | -0.408 | -0.0489 | No | ||
| 170 | EGLN1 | 2480180 3120253 6110195 | 17892 | -0.502 | -0.0398 | No | ||
| 171 | EHD4 | 5890278 | 17910 | -0.511 | -0.0179 | No | ||
| 172 | LUC7L | 6040156 6110411 | 18088 | -0.608 | -0.0005 | No | ||
| 173 | TRAM1 | 2340441 6620592 | 18164 | -0.652 | 0.0245 | No |

