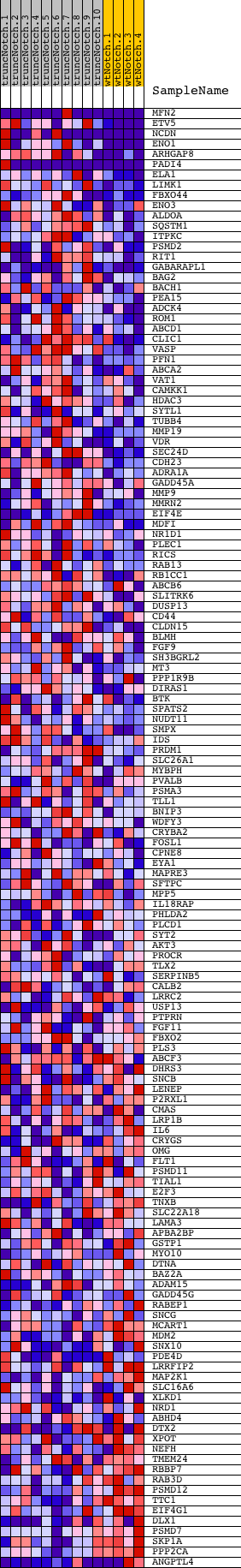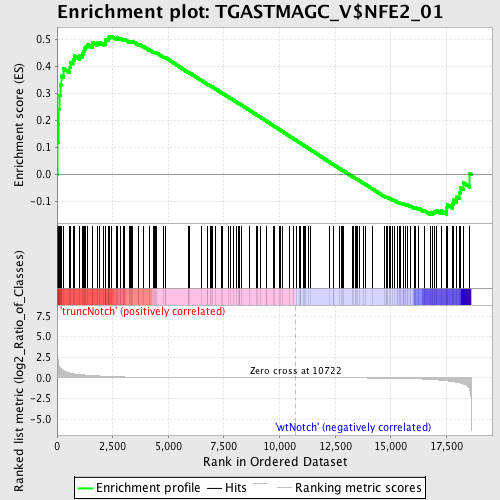
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | truncNotch |
| GeneSet | TGASTMAGC_V$NFE2_01 |
| Enrichment Score (ES) | 0.51100713 |
| Normalized Enrichment Score (NES) | 1.2740026 |
| Nominal p-value | 0.05107084 |
| FDR q-value | 0.88147247 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MFN2 | 2260195 6100164 | 12 | 3.462 | 0.1197 | Yes | ||
| 2 | ETV5 | 110017 | 54 | 1.975 | 0.1861 | Yes | ||
| 3 | NCDN | 50040 1980739 3520603 | 77 | 1.599 | 0.2405 | Yes | ||
| 4 | ENO1 | 5340128 | 90 | 1.526 | 0.2929 | Yes | ||
| 5 | ARHGAP8 | 380333 5390487 | 148 | 1.174 | 0.3306 | Yes | ||
| 6 | PADI4 | 1740075 4810441 | 204 | 1.039 | 0.3638 | Yes | ||
| 7 | ELA1 | 3390167 | 289 | 0.914 | 0.3910 | Yes | ||
| 8 | LIMK1 | 5080064 | 576 | 0.577 | 0.3956 | Yes | ||
| 9 | FBXO44 | 4760372 | 585 | 0.570 | 0.4150 | Yes | ||
| 10 | ENO3 | 5270136 | 715 | 0.501 | 0.4254 | Yes | ||
| 11 | ALDOA | 6290672 | 779 | 0.478 | 0.4386 | Yes | ||
| 12 | SQSTM1 | 6550056 | 1025 | 0.393 | 0.4390 | Yes | ||
| 13 | ITPKC | 5420021 | 1128 | 0.366 | 0.4462 | Yes | ||
| 14 | PSMD2 | 4670706 5050364 | 1184 | 0.356 | 0.4556 | Yes | ||
| 15 | RIT1 | 5390338 | 1212 | 0.352 | 0.4664 | Yes | ||
| 16 | GABARAPL1 | 2810458 | 1295 | 0.336 | 0.4736 | Yes | ||
| 17 | BAG2 | 2900324 | 1373 | 0.320 | 0.4806 | Yes | ||
| 18 | BACH1 | 290195 | 1578 | 0.281 | 0.4793 | Yes | ||
| 19 | PEA15 | 4590685 | 1580 | 0.281 | 0.4890 | Yes | ||
| 20 | ADCK4 | 1660180 | 1795 | 0.247 | 0.4860 | Yes | ||
| 21 | ROM1 | 3130685 | 1921 | 0.228 | 0.4871 | Yes | ||
| 22 | ABCD1 | 5670373 | 2091 | 0.203 | 0.4851 | Yes | ||
| 23 | CLIC1 | 110600 | 2161 | 0.194 | 0.4881 | Yes | ||
| 24 | VASP | 7050500 | 2187 | 0.191 | 0.4933 | Yes | ||
| 25 | PFN1 | 6130132 | 2194 | 0.190 | 0.4996 | Yes | ||
| 26 | ABCA2 | 5050039 | 2296 | 0.177 | 0.5003 | Yes | ||
| 27 | VAT1 | 1050040 | 2297 | 0.177 | 0.5065 | Yes | ||
| 28 | CAMKK1 | 6370324 | 2357 | 0.169 | 0.5092 | Yes | ||
| 29 | HDAC3 | 4060072 | 2427 | 0.161 | 0.5110 | Yes | ||
| 30 | SYTL1 | 510487 | 2663 | 0.136 | 0.5030 | No | ||
| 31 | TUBB4 | 5310681 | 2695 | 0.132 | 0.5059 | No | ||
| 32 | MMP19 | 5080377 | 2848 | 0.118 | 0.5018 | No | ||
| 33 | VDR | 130156 510438 | 2988 | 0.107 | 0.4980 | No | ||
| 34 | SEC24D | 3060279 | 3041 | 0.104 | 0.4988 | No | ||
| 35 | CDH23 | 4250242 5290397 6550397 | 3237 | 0.091 | 0.4914 | No | ||
| 36 | ADRA1A | 1230446 2260390 | 3302 | 0.088 | 0.4910 | No | ||
| 37 | GADD45A | 2900717 | 3324 | 0.086 | 0.4929 | No | ||
| 38 | MMP9 | 580338 | 3401 | 0.082 | 0.4916 | No | ||
| 39 | MMRN2 | 1740047 | 3670 | 0.069 | 0.4795 | No | ||
| 40 | EIF4E | 1580403 70133 6380215 | 3677 | 0.069 | 0.4816 | No | ||
| 41 | MDFI | 6200041 | 3888 | 0.061 | 0.4723 | No | ||
| 42 | NR1D1 | 2360471 770746 6590204 | 4145 | 0.053 | 0.4603 | No | ||
| 43 | PLEC1 | 6520292 50500 380075 1570088 2510692 2630497 2680017 2900746 4560497 4850376 6290398 7040471 | 4349 | 0.047 | 0.4509 | No | ||
| 44 | RICS | 1940673 | 4377 | 0.046 | 0.4511 | No | ||
| 45 | RAB13 | 3710309 | 4431 | 0.045 | 0.4498 | No | ||
| 46 | RB1CC1 | 510494 7100072 | 4445 | 0.045 | 0.4507 | No | ||
| 47 | ABCB6 | 6350138 | 4792 | 0.038 | 0.4332 | No | ||
| 48 | SLITRK6 | 7320739 | 4870 | 0.036 | 0.4303 | No | ||
| 49 | DUSP13 | 6400152 | 4891 | 0.036 | 0.4305 | No | ||
| 50 | CD44 | 3990072 4850671 5860411 6860148 7050551 | 5918 | 0.022 | 0.3757 | No | ||
| 51 | CLDN15 | 4150270 4730592 | 5922 | 0.022 | 0.3763 | No | ||
| 52 | BLMH | 1410438 | 5939 | 0.021 | 0.3762 | No | ||
| 53 | FGF9 | 1050195 | 6503 | 0.016 | 0.3463 | No | ||
| 54 | SH3BGRL2 | 1780239 | 6743 | 0.015 | 0.3338 | No | ||
| 55 | MT3 | 1450537 | 6770 | 0.015 | 0.3329 | No | ||
| 56 | PPP1R9B | 3130619 | 6884 | 0.014 | 0.3273 | No | ||
| 57 | DIRAS1 | 2640270 | 6923 | 0.014 | 0.3257 | No | ||
| 58 | BTK | 3130044 | 7004 | 0.013 | 0.3219 | No | ||
| 59 | SPATS2 | 2360066 | 7110 | 0.013 | 0.3166 | No | ||
| 60 | NUDT11 | 3840239 | 7378 | 0.011 | 0.3025 | No | ||
| 61 | SMPX | 6590440 | 7422 | 0.011 | 0.3006 | No | ||
| 62 | IDS | 360619 5720161 | 7714 | 0.010 | 0.2852 | No | ||
| 63 | PRDM1 | 3170347 3520301 | 7781 | 0.009 | 0.2819 | No | ||
| 64 | SLC26A1 | 2190142 | 7918 | 0.009 | 0.2749 | No | ||
| 65 | MYBPH | 2190711 | 8041 | 0.008 | 0.2686 | No | ||
| 66 | PVALB | 5570450 | 8167 | 0.008 | 0.2621 | No | ||
| 67 | PSMA3 | 5900047 7040161 | 8190 | 0.008 | 0.2612 | No | ||
| 68 | TLL1 | 2680315 | 8219 | 0.008 | 0.2599 | No | ||
| 69 | BNIP3 | 3140270 | 8305 | 0.007 | 0.2556 | No | ||
| 70 | WDFY3 | 3610041 4560333 4570273 | 8653 | 0.006 | 0.2370 | No | ||
| 71 | CRYBA2 | 5900138 | 8949 | 0.005 | 0.2212 | No | ||
| 72 | FOSL1 | 430021 | 8996 | 0.005 | 0.2189 | No | ||
| 73 | CPNE8 | 7040039 | 9158 | 0.004 | 0.2103 | No | ||
| 74 | EYA1 | 1450278 5220390 | 9394 | 0.004 | 0.1977 | No | ||
| 75 | MAPRE3 | 7050504 | 9403 | 0.004 | 0.1974 | No | ||
| 76 | SFTPC | 3290133 | 9725 | 0.003 | 0.1801 | No | ||
| 77 | MPP5 | 2100148 5220632 | 9790 | 0.003 | 0.1768 | No | ||
| 78 | IL18RAP | 3850088 | 10002 | 0.002 | 0.1654 | No | ||
| 79 | PHLDA2 | 4810494 | 10046 | 0.002 | 0.1632 | No | ||
| 80 | PLCD1 | 2030484 | 10136 | 0.002 | 0.1584 | No | ||
| 81 | SYT2 | 60609 | 10459 | 0.001 | 0.1410 | No | ||
| 82 | AKT3 | 1580270 3290278 | 10642 | 0.000 | 0.1311 | No | ||
| 83 | PROCR | 4920687 | 10764 | -0.000 | 0.1246 | No | ||
| 84 | TLX2 | 1090128 2900451 | 10882 | -0.000 | 0.1183 | No | ||
| 85 | SERPINB5 | 6940050 | 10942 | -0.001 | 0.1151 | No | ||
| 86 | CALB2 | 6130170 6590368 | 10951 | -0.001 | 0.1147 | No | ||
| 87 | LRRC2 | 3130400 | 11078 | -0.001 | 0.1079 | No | ||
| 88 | USP13 | 1450670 | 11099 | -0.001 | 0.1069 | No | ||
| 89 | PTPRN | 5900577 | 11167 | -0.001 | 0.1033 | No | ||
| 90 | FGF11 | 840292 | 11319 | -0.002 | 0.0952 | No | ||
| 91 | FBXO2 | 6510091 | 11368 | -0.002 | 0.0926 | No | ||
| 92 | PLS3 | 3290082 | 12242 | -0.005 | 0.0456 | No | ||
| 93 | ABCF3 | 7100100 | 12425 | -0.006 | 0.0359 | No | ||
| 94 | DHRS3 | 360609 | 12432 | -0.006 | 0.0358 | No | ||
| 95 | SNCB | 3360133 | 12674 | -0.007 | 0.0230 | No | ||
| 96 | LENEP | 4850369 | 12762 | -0.007 | 0.0185 | No | ||
| 97 | P2RXL1 | 4730390 5220446 | 12818 | -0.007 | 0.0158 | No | ||
| 98 | CMAS | 2190129 | 12857 | -0.008 | 0.0140 | No | ||
| 99 | LRP1B | 1450193 | 13290 | -0.010 | -0.0091 | No | ||
| 100 | IL6 | 380133 | 13300 | -0.010 | -0.0092 | No | ||
| 101 | CRYGS | 2680398 | 13424 | -0.011 | -0.0155 | No | ||
| 102 | OMG | 6760066 | 13470 | -0.011 | -0.0175 | No | ||
| 103 | FLT1 | 3830167 4920438 | 13493 | -0.012 | -0.0183 | No | ||
| 104 | PSMD11 | 2340538 6510053 | 13573 | -0.012 | -0.0221 | No | ||
| 105 | TIAL1 | 4150048 6510605 | 13782 | -0.014 | -0.0329 | No | ||
| 106 | E2F3 | 50162 460180 | 13841 | -0.015 | -0.0355 | No | ||
| 107 | TNXB | 630592 3830020 5360497 7000673 | 14176 | -0.019 | -0.0530 | No | ||
| 108 | SLC22A18 | 7050168 | 14706 | -0.029 | -0.0806 | No | ||
| 109 | LAMA3 | 1190131 5290239 | 14790 | -0.031 | -0.0840 | No | ||
| 110 | APBA2BP | 5700341 | 14798 | -0.031 | -0.0833 | No | ||
| 111 | GSTP1 | 3170102 3710707 5080368 | 14832 | -0.032 | -0.0840 | No | ||
| 112 | MYO10 | 3830576 5570092 | 14918 | -0.035 | -0.0874 | No | ||
| 113 | DTNA | 1340600 1780731 2340278 2850132 | 14995 | -0.037 | -0.0902 | No | ||
| 114 | BAZ2A | 730184 | 15091 | -0.041 | -0.0939 | No | ||
| 115 | ADAM15 | 5890707 | 15157 | -0.044 | -0.0959 | No | ||
| 116 | GADD45G | 2510142 | 15301 | -0.049 | -0.1020 | No | ||
| 117 | RABEP1 | 4760427 6200463 | 15393 | -0.054 | -0.1050 | No | ||
| 118 | SNCG | 3120725 | 15445 | -0.057 | -0.1058 | No | ||
| 119 | MCART1 | 5270059 | 15567 | -0.065 | -0.1101 | No | ||
| 120 | MDM2 | 3450053 5080138 | 15571 | -0.065 | -0.1080 | No | ||
| 121 | SNX10 | 3800035 | 15677 | -0.072 | -0.1112 | No | ||
| 122 | PDE4D | 2470528 6660014 | 15736 | -0.076 | -0.1117 | No | ||
| 123 | LRRFIP2 | 670575 730152 6590438 | 15880 | -0.086 | -0.1164 | No | ||
| 124 | MAP2K1 | 840739 | 16069 | -0.102 | -0.1231 | No | ||
| 125 | SLC16A6 | 1690156 | 16092 | -0.104 | -0.1207 | No | ||
| 126 | XLKD1 | 520441 | 16255 | -0.120 | -0.1253 | No | ||
| 127 | NRD1 | 5900017 | 16491 | -0.149 | -0.1328 | No | ||
| 128 | ABHD4 | 6760450 | 16760 | -0.188 | -0.1408 | No | ||
| 129 | DTX2 | 4210041 7000008 | 16891 | -0.213 | -0.1404 | No | ||
| 130 | XPOT | 7050184 | 16946 | -0.222 | -0.1356 | No | ||
| 131 | NEFH | 630239 | 17051 | -0.242 | -0.1328 | No | ||
| 132 | TMEM24 | 5860039 | 17270 | -0.295 | -0.1344 | No | ||
| 133 | RBBP7 | 430113 450450 2370309 | 17510 | -0.357 | -0.1349 | No | ||
| 134 | RAB3D | 4210253 | 17519 | -0.359 | -0.1228 | No | ||
| 135 | PSMD12 | 730044 | 17531 | -0.365 | -0.1108 | No | ||
| 136 | TTC1 | 5890736 | 17789 | -0.459 | -0.1087 | No | ||
| 137 | EIF4G1 | 4070446 | 17816 | -0.472 | -0.0937 | No | ||
| 138 | DLX1 | 360168 | 17968 | -0.540 | -0.0831 | No | ||
| 139 | PSMD7 | 2030619 6220594 | 18080 | -0.604 | -0.0681 | No | ||
| 140 | SKP1A | 2450102 | 18119 | -0.623 | -0.0485 | No | ||
| 141 | PPP2CA | 3990113 | 18255 | -0.725 | -0.0307 | No | ||
| 142 | ANGPTL4 | 2480195 6760593 | 18550 | -1.443 | 0.0036 | No |