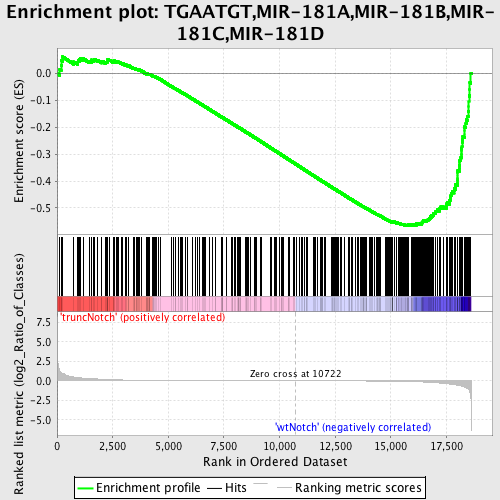
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | TGAATGT,MIR-181A,MIR-181B,MIR-181C,MIR-181D |
| Enrichment Score (ES) | -0.56728834 |
| Normalized Enrichment Score (NES) | -1.6452955 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.03251027 |
| FWER p-Value | 0.256 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TOX | 3440053 | 123 | 1.267 | 0.0155 | No | ||
| 2 | SLC38A2 | 1470242 3800026 | 180 | 1.086 | 0.0315 | No | ||
| 3 | HRB | 1660397 6110398 | 206 | 1.037 | 0.0483 | No | ||
| 4 | TGFBI | 2060446 6900112 | 250 | 0.957 | 0.0627 | No | ||
| 5 | B4GALT1 | 6980167 | 753 | 0.488 | 0.0438 | No | ||
| 6 | CARD11 | 70338 | 910 | 0.430 | 0.0428 | No | ||
| 7 | TARDBP | 870390 2350093 | 938 | 0.419 | 0.0487 | No | ||
| 8 | CHD7 | 3870372 | 998 | 0.403 | 0.0525 | No | ||
| 9 | BCL2L11 | 780044 4200601 | 1056 | 0.384 | 0.0561 | No | ||
| 10 | ETV6 | 610524 | 1176 | 0.357 | 0.0559 | No | ||
| 11 | ADM | 4810121 | 1468 | 0.301 | 0.0452 | No | ||
| 12 | SLC2A3 | 1990377 | 1539 | 0.287 | 0.0464 | No | ||
| 13 | MAP3K3 | 610685 | 1546 | 0.286 | 0.0511 | No | ||
| 14 | BHLHB2 | 7040603 | 1637 | 0.269 | 0.0509 | No | ||
| 15 | PLEKHJ1 | 4050685 | 1674 | 0.265 | 0.0536 | No | ||
| 16 | PIAS3 | 50204 240739 4060524 | 1810 | 0.244 | 0.0505 | No | ||
| 17 | AGPAT1 | 610056 | 2007 | 0.215 | 0.0436 | No | ||
| 18 | ANKRD50 | 870543 | 2013 | 0.214 | 0.0470 | No | ||
| 19 | FKBP1A | 2450368 | 2169 | 0.193 | 0.0419 | No | ||
| 20 | HYOU1 | 3060465 | 2204 | 0.189 | 0.0434 | No | ||
| 21 | ADRBK1 | 1340333 | 2249 | 0.183 | 0.0442 | No | ||
| 22 | HIC2 | 4010433 | 2259 | 0.182 | 0.0469 | No | ||
| 23 | CACNA2D2 | 520736 | 2268 | 0.181 | 0.0496 | No | ||
| 24 | TBC1D1 | 2510072 | 2277 | 0.180 | 0.0524 | No | ||
| 25 | COL16A1 | 1780520 | 2369 | 0.168 | 0.0503 | No | ||
| 26 | PAK4 | 1690152 | 2537 | 0.149 | 0.0438 | No | ||
| 27 | CAMK2G | 630097 | 2549 | 0.148 | 0.0458 | No | ||
| 28 | CRSP7 | 4230408 | 2561 | 0.147 | 0.0478 | No | ||
| 29 | MTMR9 | 2370605 | 2678 | 0.134 | 0.0438 | No | ||
| 30 | DPYSL2 | 2100427 3130112 5700324 | 2691 | 0.133 | 0.0454 | No | ||
| 31 | CRIM1 | 130491 | 2763 | 0.126 | 0.0438 | No | ||
| 32 | ESM1 | 1410594 | 2904 | 0.114 | 0.0381 | No | ||
| 33 | CYR61 | 1240408 5290026 4120452 6550008 | 2959 | 0.109 | 0.0371 | No | ||
| 34 | EIF4A2 | 1170494 1740711 2850504 | 3059 | 0.103 | 0.0335 | No | ||
| 35 | LRP12 | 2370112 | 3109 | 0.100 | 0.0325 | No | ||
| 36 | MAP3K10 | 3360168 4850180 | 3211 | 0.093 | 0.0286 | No | ||
| 37 | CPNE2 | 60551 | 3216 | 0.093 | 0.0301 | No | ||
| 38 | ETS1 | 5270278 6450717 6620465 | 3446 | 0.079 | 0.0189 | No | ||
| 39 | RGMA | 2450711 | 3465 | 0.079 | 0.0193 | No | ||
| 40 | PCDHA11 | 1510253 | 3488 | 0.078 | 0.0195 | No | ||
| 41 | EGR3 | 6940128 | 3572 | 0.073 | 0.0162 | No | ||
| 42 | PCNP | 610768 | 3575 | 0.073 | 0.0174 | No | ||
| 43 | MIP | 6520333 | 3606 | 0.072 | 0.0170 | No | ||
| 44 | EPS15 | 4200215 | 3674 | 0.069 | 0.0146 | No | ||
| 45 | ATXN1 | 5550156 | 3707 | 0.068 | 0.0140 | No | ||
| 46 | FOS | 1850315 | 3778 | 0.065 | 0.0113 | No | ||
| 47 | GALNTL1 | 2850497 | 4005 | 0.058 | -0.0000 | No | ||
| 48 | DOCK10 | 5890402 6200279 6770025 | 4036 | 0.056 | -0.0007 | No | ||
| 49 | MAP1A | 4920576 | 4047 | 0.056 | -0.0003 | No | ||
| 50 | PPP1CB | 3130411 3780270 4150180 | 4083 | 0.055 | -0.0012 | No | ||
| 51 | CSNK1G1 | 840082 1230575 3940647 | 4124 | 0.053 | -0.0025 | No | ||
| 52 | UNC5A | 2120528 3060324 | 4149 | 0.053 | -0.0029 | No | ||
| 53 | TBC1D4 | 6420397 | 4151 | 0.053 | -0.0020 | No | ||
| 54 | SPRY4 | 1570594 | 4169 | 0.052 | -0.0020 | No | ||
| 55 | CDYL | 4200142 4730397 6060400 | 4294 | 0.048 | -0.0079 | No | ||
| 56 | ZBTB4 | 6450441 | 4331 | 0.047 | -0.0091 | No | ||
| 57 | PHLDA1 | 2450020 | 4354 | 0.047 | -0.0095 | No | ||
| 58 | KLHL5 | 3060484 6400292 | 4381 | 0.046 | -0.0101 | No | ||
| 59 | SOX5 | 2370576 2900167 3190128 5050528 | 4427 | 0.045 | -0.0117 | No | ||
| 60 | KLHL2 | 360551 | 4458 | 0.045 | -0.0126 | No | ||
| 61 | PCDHA5 | 2970347 | 4570 | 0.042 | -0.0179 | No | ||
| 62 | GGTL3 | 1770288 | 4665 | 0.040 | -0.0224 | No | ||
| 63 | PER2 | 3120204 3390593 | 5139 | 0.032 | -0.0477 | No | ||
| 64 | BACH2 | 6760131 | 5241 | 0.030 | -0.0527 | No | ||
| 65 | SYPL2 | 2450113 | 5316 | 0.029 | -0.0562 | No | ||
| 66 | LIF | 1410373 2060129 3990338 | 5327 | 0.029 | -0.0563 | No | ||
| 67 | HOXC8 | 6220050 | 5477 | 0.027 | -0.0640 | No | ||
| 68 | PCDHA6 | 780333 2370280 | 5532 | 0.026 | -0.0665 | No | ||
| 69 | CDH13 | 5130368 5340114 | 5585 | 0.025 | -0.0689 | No | ||
| 70 | ROBO2 | 450136 | 5655 | 0.025 | -0.0722 | No | ||
| 71 | PKNOX2 | 610066 2810075 4230601 | 5776 | 0.023 | -0.0784 | No | ||
| 72 | BMPR2 | 5340066 6960328 | 5843 | 0.022 | -0.0816 | No | ||
| 73 | CPD | 5360403 5860685 | 5864 | 0.022 | -0.0823 | No | ||
| 74 | IL1A | 3610056 | 6101 | 0.020 | -0.0948 | No | ||
| 75 | SYT3 | 6380017 | 6216 | 0.019 | -0.1007 | No | ||
| 76 | SIX2 | 1450551 | 6301 | 0.018 | -0.1050 | No | ||
| 77 | HOXB5 | 2900538 | 6404 | 0.017 | -0.1103 | No | ||
| 78 | GRM5 | 60528 | 6513 | 0.016 | -0.1159 | No | ||
| 79 | CUL5 | 450142 | 6562 | 0.016 | -0.1183 | No | ||
| 80 | STAG1 | 1190300 1400722 4590100 | 6644 | 0.015 | -0.1224 | No | ||
| 81 | CNR1 | 6550494 6760497 | 6673 | 0.015 | -0.1237 | No | ||
| 82 | CNTNAP2 | 360170 4280546 | 6828 | 0.014 | -0.1319 | No | ||
| 83 | BCL11A | 6860369 | 6996 | 0.013 | -0.1408 | No | ||
| 84 | SYNPR | 2810400 | 7113 | 0.013 | -0.1469 | No | ||
| 85 | OSBPL8 | 50603 | 7136 | 0.012 | -0.1479 | No | ||
| 86 | KLF15 | 4230164 | 7379 | 0.011 | -0.1609 | No | ||
| 87 | SOX6 | 2940458 4070601 6840717 | 7381 | 0.011 | -0.1608 | No | ||
| 88 | PAK7 | 3830164 | 7423 | 0.011 | -0.1628 | No | ||
| 89 | TRIM9 | 1230672 2120162 6450100 6450687 | 7424 | 0.011 | -0.1626 | No | ||
| 90 | TNFRSF11B | 4610446 | 7425 | 0.011 | -0.1624 | No | ||
| 91 | BIRC6 | 70148 | 7596 | 0.010 | -0.1716 | No | ||
| 92 | CDON | 540204 | 7629 | 0.010 | -0.1731 | No | ||
| 93 | ARHGEF7 | 1690441 | 7829 | 0.009 | -0.1838 | No | ||
| 94 | NPTXR | 4060685 450438 | 7884 | 0.009 | -0.1866 | No | ||
| 95 | ENAH | 1690292 5700300 | 7886 | 0.009 | -0.1865 | No | ||
| 96 | RKHD3 | 6180471 | 7952 | 0.009 | -0.1899 | No | ||
| 97 | HLF | 2370113 | 8006 | 0.008 | -0.1927 | No | ||
| 98 | SUHW4 | 7000075 | 8121 | 0.008 | -0.1988 | No | ||
| 99 | DOCK7 | 1780398 | 8130 | 0.008 | -0.1991 | No | ||
| 100 | PCDHA12 | 2810215 | 8164 | 0.008 | -0.2008 | No | ||
| 101 | LPP | 1660541 | 8169 | 0.008 | -0.2008 | No | ||
| 102 | IGSF11 | 60471 2350288 | 8184 | 0.008 | -0.2015 | No | ||
| 103 | SEPT3 | 3830022 5290079 | 8222 | 0.008 | -0.2034 | No | ||
| 104 | CBLB | 3520465 4670348 | 8240 | 0.008 | -0.2042 | No | ||
| 105 | MAMDC2 | 430471 | 8447 | 0.007 | -0.2153 | No | ||
| 106 | ADARB1 | 780451 | 8492 | 0.007 | -0.2176 | No | ||
| 107 | PHOX2B | 5270075 | 8527 | 0.006 | -0.2193 | No | ||
| 108 | BAI3 | 940692 | 8550 | 0.006 | -0.2204 | No | ||
| 109 | FIGN | 7000601 | 8600 | 0.006 | -0.2230 | No | ||
| 110 | PRKCE | 5700053 | 8671 | 0.006 | -0.2267 | No | ||
| 111 | HOXA11 | 6980133 | 8875 | 0.005 | -0.2377 | No | ||
| 112 | CDC42BPA | 840671 | 8877 | 0.005 | -0.2377 | No | ||
| 113 | GREM1 | 3940180 | 8905 | 0.005 | -0.2391 | No | ||
| 114 | PCDHA10 | 3830725 | 8970 | 0.005 | -0.2425 | No | ||
| 115 | TRIM2 | 430546 1170286 | 9142 | 0.005 | -0.2518 | No | ||
| 116 | ACVR2A | 6110647 | 9155 | 0.004 | -0.2524 | No | ||
| 117 | ADAMTS1 | 3140286 5860463 | 9197 | 0.004 | -0.2545 | No | ||
| 118 | RAB11FIP2 | 4570040 | 9569 | 0.003 | -0.2748 | No | ||
| 119 | KLF6 | 110541 5360026 | 9655 | 0.003 | -0.2793 | No | ||
| 120 | MPP5 | 2100148 5220632 | 9790 | 0.003 | -0.2866 | No | ||
| 121 | LRRN1 | 3290154 | 9805 | 0.003 | -0.2874 | No | ||
| 122 | GRB10 | 6980082 | 9840 | 0.002 | -0.2892 | No | ||
| 123 | BTBD3 | 1980041 2510577 3190053 4200746 | 9988 | 0.002 | -0.2972 | No | ||
| 124 | SCHIP1 | 4560152 | 10093 | 0.002 | -0.3028 | No | ||
| 125 | ESR1 | 4060372 5860193 | 10107 | 0.002 | -0.3035 | No | ||
| 126 | TOM1L1 | 4920056 3190070 | 10128 | 0.002 | -0.3046 | No | ||
| 127 | RPS6KB1 | 1450427 5080110 6200563 | 10168 | 0.002 | -0.3067 | No | ||
| 128 | NR2C2 | 6100097 | 10395 | 0.001 | -0.3190 | No | ||
| 129 | CD163 | 2480315 | 10415 | 0.001 | -0.3201 | No | ||
| 130 | PDGFRA | 2940332 | 10436 | 0.001 | -0.3211 | No | ||
| 131 | AKT3 | 1580270 3290278 | 10642 | 0.000 | -0.3323 | No | ||
| 132 | ZFP36L1 | 2510138 4120048 | 10657 | 0.000 | -0.3331 | No | ||
| 133 | NMT2 | 6900148 | 10757 | -0.000 | -0.3385 | No | ||
| 134 | IMPG1 | 5570025 | 10899 | -0.000 | -0.3462 | No | ||
| 135 | WDR37 | 2630519 6840347 | 10995 | -0.001 | -0.3514 | No | ||
| 136 | ATG5 | 6200433 5360324 | 11031 | -0.001 | -0.3533 | No | ||
| 137 | BAAT | 730739 | 11098 | -0.001 | -0.3569 | No | ||
| 138 | ARNT2 | 1740594 | 11133 | -0.001 | -0.3587 | No | ||
| 139 | SMAD7 | 430377 | 11202 | -0.001 | -0.3624 | No | ||
| 140 | KCNMA1 | 1450402 | 11220 | -0.001 | -0.3633 | No | ||
| 141 | CNTN4 | 1780300 5570577 6370019 | 11263 | -0.002 | -0.3656 | No | ||
| 142 | ZIC3 | 380020 | 11531 | -0.002 | -0.3802 | No | ||
| 143 | SSX2IP | 4610176 | 11579 | -0.002 | -0.3827 | No | ||
| 144 | ADAMTS18 | 5690008 | 11619 | -0.003 | -0.3848 | No | ||
| 145 | PLAG1 | 1450142 3870139 6520039 | 11708 | -0.003 | -0.3895 | No | ||
| 146 | RAN | 2260446 4590647 | 11821 | -0.003 | -0.3956 | No | ||
| 147 | PAM | 5290528 | 11858 | -0.003 | -0.3975 | No | ||
| 148 | RBM22 | 5570064 | 11875 | -0.003 | -0.3983 | No | ||
| 149 | GABRA1 | 130372 | 11904 | -0.004 | -0.3998 | No | ||
| 150 | CCNJ | 870047 | 11934 | -0.004 | -0.4013 | No | ||
| 151 | ZIC2 | 1410408 | 11940 | -0.004 | -0.4015 | No | ||
| 152 | GRIK2 | 4610164 | 11999 | -0.004 | -0.4046 | No | ||
| 153 | NPEPPS | 2630731 | 12053 | -0.004 | -0.4075 | No | ||
| 154 | E2F5 | 5860575 | 12350 | -0.005 | -0.4236 | No | ||
| 155 | SLC25A37 | 6520341 460471 | 12360 | -0.005 | -0.4240 | No | ||
| 156 | THRB | 1780673 | 12363 | -0.005 | -0.4240 | No | ||
| 157 | EPHA4 | 460750 | 12408 | -0.005 | -0.4263 | No | ||
| 158 | UBE2B | 4780497 | 12466 | -0.006 | -0.4293 | No | ||
| 159 | PLCL2 | 6100575 | 12478 | -0.006 | -0.4298 | No | ||
| 160 | TNFSF11 | 6350204 | 12531 | -0.006 | -0.4325 | No | ||
| 161 | PCDHA3 | 3840402 | 12568 | -0.006 | -0.4344 | No | ||
| 162 | CBX4 | 70332 | 12623 | -0.006 | -0.4373 | No | ||
| 163 | TBPL1 | 3610332 | 12665 | -0.007 | -0.4394 | No | ||
| 164 | DPP10 | 4730746 | 12732 | -0.007 | -0.4429 | No | ||
| 165 | LIN28 | 6590672 | 12789 | -0.007 | -0.4458 | No | ||
| 166 | E2F7 | 4200056 | 12926 | -0.008 | -0.4531 | No | ||
| 167 | NEGR1 | 1940731 5130170 | 13107 | -0.009 | -0.4628 | No | ||
| 168 | ATP2B2 | 3780397 | 13136 | -0.009 | -0.4642 | No | ||
| 169 | SLITRK2 | 2640020 | 13159 | -0.009 | -0.4652 | No | ||
| 170 | CCNK | 6980048 | 13233 | -0.010 | -0.4690 | No | ||
| 171 | LMO1 | 3170528 | 13253 | -0.010 | -0.4699 | No | ||
| 172 | TCERG1 | 2350551 4540397 5220746 | 13257 | -0.010 | -0.4699 | No | ||
| 173 | SEMA4C | 1980315 | 13389 | -0.011 | -0.4769 | No | ||
| 174 | OSBPL3 | 3170164 4210196 | 13480 | -0.012 | -0.4816 | No | ||
| 175 | USP33 | 780047 | 13559 | -0.012 | -0.4856 | No | ||
| 176 | DDX3Y | 1580278 4200519 | 13631 | -0.013 | -0.4893 | No | ||
| 177 | PCDHA1 | 940026 540484 | 13688 | -0.013 | -0.4921 | No | ||
| 178 | HECA | 3310612 | 13709 | -0.013 | -0.4930 | No | ||
| 179 | ST8SIA4 | 2680605 3060215 | 13750 | -0.014 | -0.4949 | No | ||
| 180 | GRIA2 | 5080088 | 13763 | -0.014 | -0.4953 | No | ||
| 181 | EN2 | 7000368 | 13785 | -0.014 | -0.4962 | No | ||
| 182 | UBP1 | 6020707 | 13803 | -0.014 | -0.4969 | No | ||
| 183 | PBX3 | 1300424 3710577 6180575 | 13851 | -0.015 | -0.4992 | No | ||
| 184 | PDK4 | 6400300 | 13905 | -0.015 | -0.5019 | No | ||
| 185 | HMGB2 | 2640603 | 13918 | -0.016 | -0.5022 | No | ||
| 186 | RSBN1 | 7000487 | 13919 | -0.016 | -0.5020 | No | ||
| 187 | ANKRD13C | 5270202 | 14052 | -0.017 | -0.5089 | No | ||
| 188 | NRXN1 | 460408 5340270 6370066 | 14072 | -0.017 | -0.5096 | No | ||
| 189 | PCGF2 | 6370347 | 14132 | -0.018 | -0.5125 | No | ||
| 190 | PALM2 | 4120068 7100398 | 14167 | -0.019 | -0.5141 | No | ||
| 191 | SPIRE1 | 610286 2230142 3120154 | 14281 | -0.020 | -0.5199 | No | ||
| 192 | NR6A1 | 4010347 | 14357 | -0.022 | -0.5236 | No | ||
| 193 | NCOA2 | 940347 2850725 | 14393 | -0.022 | -0.5251 | No | ||
| 194 | TRIM3 | 5570156 | 14403 | -0.022 | -0.5252 | No | ||
| 195 | MINK1 | 5860647 | 14444 | -0.023 | -0.5270 | No | ||
| 196 | NR4A3 | 2900021 5860095 5910039 | 14482 | -0.024 | -0.5286 | No | ||
| 197 | QKI | 5220093 6130044 | 14486 | -0.024 | -0.5284 | No | ||
| 198 | SLC25A25 | 1230605 | 14513 | -0.024 | -0.5294 | No | ||
| 199 | ATP11C | 770672 7040129 | 14555 | -0.025 | -0.5312 | No | ||
| 200 | WDR20 | 1240373 | 14754 | -0.030 | -0.5415 | No | ||
| 201 | SEMA4G | 5890026 | 14791 | -0.031 | -0.5429 | No | ||
| 202 | ZDHHC7 | 1500014 | 14846 | -0.032 | -0.5453 | No | ||
| 203 | BAIAP2 | 460347 4730600 | 14877 | -0.033 | -0.5463 | No | ||
| 204 | TRAK2 | 430736 1770026 | 14936 | -0.035 | -0.5489 | No | ||
| 205 | SLC9A6 | 4570091 7100097 | 14975 | -0.036 | -0.5503 | No | ||
| 206 | CLASP1 | 6860279 | 15001 | -0.037 | -0.5511 | No | ||
| 207 | PTPN9 | 3290408 | 15031 | -0.039 | -0.5520 | No | ||
| 208 | ARRDC3 | 4610390 | 15053 | -0.039 | -0.5524 | No | ||
| 209 | NLK | 2030010 2450041 | 15071 | -0.040 | -0.5527 | No | ||
| 210 | CAMSAP1 | 1580131 2120494 6620446 | 15073 | -0.040 | -0.5520 | No | ||
| 211 | RECK | 840079 | 15081 | -0.040 | -0.5517 | No | ||
| 212 | FNDC3A | 2690097 3290044 | 15085 | -0.040 | -0.5511 | No | ||
| 213 | GATA6 | 1770010 | 15086 | -0.041 | -0.5504 | No | ||
| 214 | BAZ2A | 730184 | 15091 | -0.041 | -0.5499 | No | ||
| 215 | PICALM | 1940280 | 15092 | -0.041 | -0.5492 | No | ||
| 216 | STC1 | 360161 | 15152 | -0.043 | -0.5517 | No | ||
| 217 | HCN2 | 3840181 | 15243 | -0.047 | -0.5558 | No | ||
| 218 | CHD9 | 430037 2570129 5420398 | 15256 | -0.048 | -0.5556 | No | ||
| 219 | SS18L1 | 4610180 6550068 | 15269 | -0.048 | -0.5554 | No | ||
| 220 | LIN7C | 4850341 | 15272 | -0.048 | -0.5547 | No | ||
| 221 | PSAP | 2810338 | 15355 | -0.052 | -0.5582 | No | ||
| 222 | ADAM11 | 1050008 3130494 | 15357 | -0.052 | -0.5574 | No | ||
| 223 | PAWR | 5130546 6330706 | 15404 | -0.055 | -0.5589 | No | ||
| 224 | ADCY9 | 5690168 | 15419 | -0.055 | -0.5587 | No | ||
| 225 | EYA3 | 1580195 6510632 | 15492 | -0.060 | -0.5616 | No | ||
| 226 | INPP5E | 6100619 | 15499 | -0.060 | -0.5609 | No | ||
| 227 | BCLAF1 | 2030239 5720278 | 15535 | -0.063 | -0.5617 | No | ||
| 228 | CREB1 | 1500717 2230358 3610600 6550601 | 15564 | -0.065 | -0.5621 | No | ||
| 229 | INPP5A | 5910195 | 15624 | -0.068 | -0.5642 | No | ||
| 230 | EN1 | 5360168 7050025 | 15637 | -0.069 | -0.5636 | No | ||
| 231 | FOXK1 | 2640373 3290136 | 15671 | -0.071 | -0.5642 | No | ||
| 232 | YTHDF3 | 4540632 4670717 7040609 | 15687 | -0.073 | -0.5637 | No | ||
| 233 | TIMP3 | 1450504 1980270 | 15744 | -0.077 | -0.5654 | No | ||
| 234 | PNRC2 | 3360309 | 15763 | -0.078 | -0.5650 | No | ||
| 235 | SLC25A3 | 670112 | 15766 | -0.079 | -0.5638 | No | ||
| 236 | PI4K2B | 2480215 4610563 | 15795 | -0.080 | -0.5639 | No | ||
| 237 | NARG1 | 5910563 6350095 | 15804 | -0.081 | -0.5629 | No | ||
| 238 | NR3C1 | 630563 | 15808 | -0.081 | -0.5617 | No | ||
| 239 | CACNB2 | 1500095 7330707 | 15912 | -0.089 | -0.5657 | Yes | ||
| 240 | ARFGEF2 | 5550500 | 15917 | -0.089 | -0.5644 | Yes | ||
| 241 | PHF15 | 6550066 6860673 | 15924 | -0.090 | -0.5631 | Yes | ||
| 242 | ROD1 | 2060324 3140332 | 15973 | -0.094 | -0.5641 | Yes | ||
| 243 | CARM1 | 7040292 | 15993 | -0.096 | -0.5635 | Yes | ||
| 244 | MAPT | 360706 1230706 | 16010 | -0.097 | -0.5627 | Yes | ||
| 245 | MAP2K1 | 840739 | 16069 | -0.102 | -0.5641 | Yes | ||
| 246 | FOXP1 | 6510433 | 16107 | -0.105 | -0.5642 | Yes | ||
| 247 | MYH10 | 2640673 | 16127 | -0.107 | -0.5634 | Yes | ||
| 248 | PCDHA7 | 1850019 | 16139 | -0.108 | -0.5621 | Yes | ||
| 249 | PUM1 | 6130500 | 16146 | -0.109 | -0.5605 | Yes | ||
| 250 | MAPK1 | 3190193 6200253 | 16153 | -0.109 | -0.5589 | Yes | ||
| 251 | FBXO33 | 1510504 | 16211 | -0.115 | -0.5600 | Yes | ||
| 252 | NAT5 | 1690044 4850242 6110427 | 16237 | -0.118 | -0.5593 | Yes | ||
| 253 | CAMTA2 | 3990075 | 16303 | -0.125 | -0.5607 | Yes | ||
| 254 | ITGA3 | 4570427 | 16313 | -0.127 | -0.5590 | Yes | ||
| 255 | SCOC | 610048 2230053 | 16369 | -0.133 | -0.5597 | Yes | ||
| 256 | DYNC1LI2 | 50338 | 16374 | -0.134 | -0.5575 | Yes | ||
| 257 | PAPOLG | 2470400 | 16394 | -0.135 | -0.5562 | Yes | ||
| 258 | PURB | 5360138 | 16402 | -0.136 | -0.5542 | Yes | ||
| 259 | NFAT5 | 2510411 5890195 6550152 | 16428 | -0.140 | -0.5531 | Yes | ||
| 260 | PCAF | 2230161 2570369 6550451 | 16443 | -0.142 | -0.5514 | Yes | ||
| 261 | ZC3H6 | 130711 | 16450 | -0.143 | -0.5492 | Yes | ||
| 262 | RTF1 | 2100735 | 16452 | -0.143 | -0.5468 | Yes | ||
| 263 | RAD21 | 1990278 | 16495 | -0.150 | -0.5464 | Yes | ||
| 264 | RPS6KA3 | 1980707 | 16549 | -0.158 | -0.5466 | Yes | ||
| 265 | POM121 | 70152 | 16607 | -0.167 | -0.5468 | Yes | ||
| 266 | DCUN1D1 | 2360332 | 16631 | -0.171 | -0.5450 | Yes | ||
| 267 | NCALD | 730176 | 16677 | -0.177 | -0.5444 | Yes | ||
| 268 | ACSL1 | 2900520 | 16682 | -0.177 | -0.5415 | Yes | ||
| 269 | TBL1X | 6400524 | 16735 | -0.184 | -0.5411 | Yes | ||
| 270 | PDIA6 | 2760632 | 16737 | -0.185 | -0.5379 | Yes | ||
| 271 | DNAJC13 | 4730091 | 16770 | -0.190 | -0.5363 | Yes | ||
| 272 | LRBA | 3450402 | 16772 | -0.191 | -0.5331 | Yes | ||
| 273 | FMNL2 | 4060438 | 16817 | -0.199 | -0.5320 | Yes | ||
| 274 | PLEKHA3 | 5080068 | 16827 | -0.201 | -0.5289 | Yes | ||
| 275 | SIRT1 | 1190731 | 16889 | -0.212 | -0.5285 | Yes | ||
| 276 | CDC73 | 1580132 | 16908 | -0.216 | -0.5257 | Yes | ||
| 277 | SEC24C | 6980707 | 16921 | -0.218 | -0.5226 | Yes | ||
| 278 | RAB11A | 5360079 | 16932 | -0.220 | -0.5193 | Yes | ||
| 279 | GOLGA1 | 5860471 | 16988 | -0.229 | -0.5183 | Yes | ||
| 280 | MAT2A | 4070026 4730079 6020280 | 17019 | -0.235 | -0.5158 | Yes | ||
| 281 | RNMT | 1770500 | 17022 | -0.236 | -0.5118 | Yes | ||
| 282 | BRWD1 | 3130048 3140168 6400601 | 17081 | -0.249 | -0.5106 | Yes | ||
| 283 | CBX7 | 3940035 | 17103 | -0.253 | -0.5073 | Yes | ||
| 284 | YTHDF2 | 4060332 4610279 | 17187 | -0.271 | -0.5071 | Yes | ||
| 285 | MKNK2 | 1240020 5270092 | 17203 | -0.275 | -0.5031 | Yes | ||
| 286 | RKHD2 | 50035 3930300 | 17216 | -0.280 | -0.4988 | Yes | ||
| 287 | MECP2 | 1940450 | 17253 | -0.290 | -0.4957 | Yes | ||
| 288 | AP1G1 | 2360671 | 17370 | -0.324 | -0.4964 | Yes | ||
| 289 | SENP1 | 6100537 | 17489 | -0.353 | -0.4967 | Yes | ||
| 290 | SFRS7 | 2760408 | 17496 | -0.354 | -0.4908 | Yes | ||
| 291 | RBBP7 | 430113 450450 2370309 | 17510 | -0.357 | -0.4853 | Yes | ||
| 292 | CSNK1G3 | 110450 | 17530 | -0.364 | -0.4799 | Yes | ||
| 293 | SENP2 | 4540452 | 17642 | -0.407 | -0.4789 | Yes | ||
| 294 | YWHAG | 3780341 | 17650 | -0.409 | -0.4721 | Yes | ||
| 295 | DAZAP2 | 6200102 | 17667 | -0.415 | -0.4657 | Yes | ||
| 296 | DDX3X | 2190020 | 17682 | -0.421 | -0.4590 | Yes | ||
| 297 | PRKCD | 770592 | 17700 | -0.425 | -0.4525 | Yes | ||
| 298 | EPC2 | 2470095 | 17729 | -0.437 | -0.4464 | Yes | ||
| 299 | CTDSPL | 4670546 | 17755 | -0.447 | -0.4399 | Yes | ||
| 300 | MMP14 | 670079 | 17840 | -0.481 | -0.4361 | Yes | ||
| 301 | ATP1B1 | 3130594 | 17850 | -0.483 | -0.4281 | Yes | ||
| 302 | MBNL2 | 3450707 | 17905 | -0.509 | -0.4221 | Yes | ||
| 303 | RANBP5 | 3780019 | 17916 | -0.514 | -0.4137 | Yes | ||
| 304 | RABGEF1 | 4010497 | 17990 | -0.551 | -0.4080 | Yes | ||
| 305 | MTMR12 | 7000181 | 17999 | -0.557 | -0.3987 | Yes | ||
| 306 | CGGBP1 | 4810053 | 18005 | -0.559 | -0.3892 | Yes | ||
| 307 | GPIAP1 | 780551 6420079 | 18008 | -0.561 | -0.3794 | Yes | ||
| 308 | CD4 | 1090010 | 18009 | -0.561 | -0.3696 | Yes | ||
| 309 | RUNX1 | 3840711 | 18014 | -0.563 | -0.3600 | Yes | ||
| 310 | KPNB1 | 1690138 | 18073 | -0.602 | -0.3526 | Yes | ||
| 311 | IVNS1ABP | 4760601 6520113 | 18081 | -0.605 | -0.3424 | Yes | ||
| 312 | KPNA1 | 5270324 | 18099 | -0.612 | -0.3326 | Yes | ||
| 313 | MTPN | 3940167 | 18102 | -0.614 | -0.3219 | Yes | ||
| 314 | METAP1 | 3520632 | 18148 | -0.642 | -0.3131 | Yes | ||
| 315 | OGT | 2360131 4610333 | 18167 | -0.653 | -0.3027 | Yes | ||
| 316 | RAP1B | 3710138 | 18172 | -0.654 | -0.2914 | Yes | ||
| 317 | UBE3C | 3140673 | 18174 | -0.656 | -0.2800 | Yes | ||
| 318 | EED | 2320373 | 18198 | -0.671 | -0.2694 | Yes | ||
| 319 | RNF6 | 2640035 | 18203 | -0.674 | -0.2578 | Yes | ||
| 320 | WSB1 | 870563 | 18210 | -0.681 | -0.2462 | Yes | ||
| 321 | NSMAF | 3870725 | 18239 | -0.703 | -0.2354 | Yes | ||
| 322 | ETF1 | 6770075 | 18309 | -0.780 | -0.2256 | Yes | ||
| 323 | PPP3R1 | 1190041 1570487 | 18310 | -0.780 | -0.2119 | Yes | ||
| 324 | SRPK2 | 6380341 | 18312 | -0.780 | -0.1983 | Yes | ||
| 325 | BRD1 | 1660278 | 18375 | -0.856 | -0.1866 | Yes | ||
| 326 | LBR | 4540671 | 18390 | -0.872 | -0.1721 | Yes | ||
| 327 | PIK3R3 | 1770333 | 18458 | -1.005 | -0.1582 | Yes | ||
| 328 | RNF34 | 3800332 4590717 5670138 | 18479 | -1.062 | -0.1406 | Yes | ||
| 329 | GHITM | 3290092 | 18510 | -1.144 | -0.1222 | Yes | ||
| 330 | CCAR1 | 3190092 | 18512 | -1.150 | -0.1021 | Yes | ||
| 331 | CUL3 | 1850520 | 18514 | -1.160 | -0.0818 | Yes | ||
| 332 | TULP4 | 2320364 | 18549 | -1.415 | -0.0589 | Yes | ||
| 333 | ATP2A2 | 1090075 3990279 | 18554 | -1.475 | -0.0332 | Yes | ||
| 334 | ILF3 | 940722 3190647 6520110 | 18599 | -2.087 | 0.0009 | Yes |

