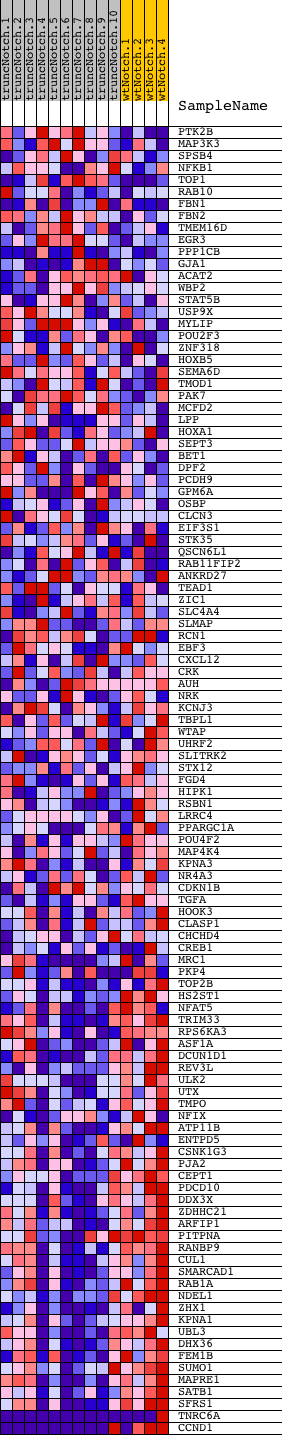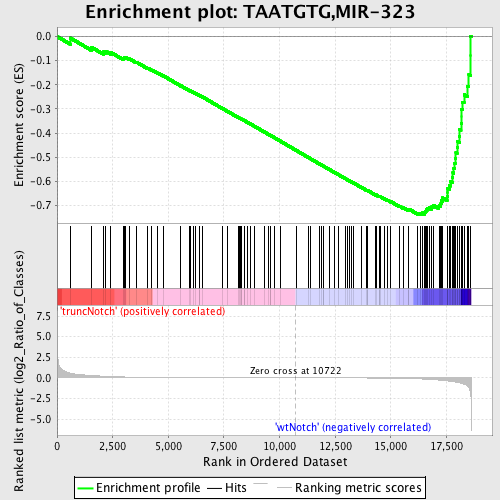
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | TAATGTG,MIR-323 |
| Enrichment Score (ES) | -0.73762476 |
| Normalized Enrichment Score (NES) | -1.9098186 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PTK2B | 4730411 | 591 | 0.567 | -0.0062 | No | ||
| 2 | MAP3K3 | 610685 | 1546 | 0.286 | -0.0447 | No | ||
| 3 | SPSB4 | 4070673 | 2076 | 0.205 | -0.0640 | No | ||
| 4 | NFKB1 | 5420358 | 2171 | 0.193 | -0.0603 | No | ||
| 5 | TOP1 | 770471 4060632 6650324 | 2420 | 0.162 | -0.0664 | No | ||
| 6 | RAB10 | 2360008 5890020 | 2970 | 0.108 | -0.0911 | No | ||
| 7 | FBN1 | 3170181 | 3009 | 0.106 | -0.0884 | No | ||
| 8 | FBN2 | 50435 | 3084 | 0.101 | -0.0877 | No | ||
| 9 | TMEM16D | 2650324 | 3255 | 0.090 | -0.0928 | No | ||
| 10 | EGR3 | 6940128 | 3572 | 0.073 | -0.1066 | No | ||
| 11 | PPP1CB | 3130411 3780270 4150180 | 4083 | 0.055 | -0.1316 | No | ||
| 12 | GJA1 | 5220731 | 4258 | 0.049 | -0.1388 | No | ||
| 13 | ACAT2 | 2680400 3610440 | 4527 | 0.043 | -0.1513 | No | ||
| 14 | WBP2 | 3170128 | 4759 | 0.038 | -0.1621 | No | ||
| 15 | STAT5B | 6200026 | 5565 | 0.026 | -0.2044 | No | ||
| 16 | USP9X | 130435 2370735 3120338 | 5965 | 0.021 | -0.2250 | No | ||
| 17 | MYLIP | 50717 4050672 | 5987 | 0.021 | -0.2252 | No | ||
| 18 | POU2F3 | 2030601 | 6150 | 0.019 | -0.2331 | No | ||
| 19 | ZNF318 | 5890091 6130021 | 6233 | 0.019 | -0.2366 | No | ||
| 20 | HOXB5 | 2900538 | 6404 | 0.017 | -0.2450 | No | ||
| 21 | SEMA6D | 4050324 5860138 6350307 | 6411 | 0.017 | -0.2446 | No | ||
| 22 | TMOD1 | 3850100 | 6534 | 0.016 | -0.2504 | No | ||
| 23 | PAK7 | 3830164 | 7423 | 0.011 | -0.2979 | No | ||
| 24 | MCFD2 | 520576 5690017 | 7654 | 0.010 | -0.3099 | No | ||
| 25 | LPP | 1660541 | 8169 | 0.008 | -0.3373 | No | ||
| 26 | HOXA1 | 1190524 5420142 | 8216 | 0.008 | -0.3395 | No | ||
| 27 | SEPT3 | 3830022 5290079 | 8222 | 0.008 | -0.3394 | No | ||
| 28 | BET1 | 4010725 | 8281 | 0.007 | -0.3422 | No | ||
| 29 | DPF2 | 6980373 | 8302 | 0.007 | -0.3429 | No | ||
| 30 | PCDH9 | 2100136 | 8403 | 0.007 | -0.3480 | No | ||
| 31 | GPM6A | 1660044 2750152 | 8430 | 0.007 | -0.3491 | No | ||
| 32 | OSBP | 5390471 | 8579 | 0.006 | -0.3568 | No | ||
| 33 | CLCN3 | 510195 780176 1660093 3830372 | 8681 | 0.006 | -0.3620 | No | ||
| 34 | EIF3S1 | 6130368 6770044 | 8891 | 0.005 | -0.3731 | No | ||
| 35 | STK35 | 2360121 | 9313 | 0.004 | -0.3956 | No | ||
| 36 | QSCN6L1 | 870040 | 9507 | 0.003 | -0.4059 | No | ||
| 37 | RAB11FIP2 | 4570040 | 9569 | 0.003 | -0.4091 | No | ||
| 38 | ANKRD27 | 2190465 4070300 6040026 | 9601 | 0.003 | -0.4106 | No | ||
| 39 | TEAD1 | 2470551 | 9756 | 0.003 | -0.4188 | No | ||
| 40 | ZIC1 | 670113 | 10040 | 0.002 | -0.4340 | No | ||
| 41 | SLC4A4 | 1780075 1850040 4810068 | 10740 | -0.000 | -0.4718 | No | ||
| 42 | SLMAP | 3120093 4760128 5390528 | 11296 | -0.002 | -0.5017 | No | ||
| 43 | RCN1 | 2480041 | 11389 | -0.002 | -0.5066 | No | ||
| 44 | EBF3 | 4200129 | 11771 | -0.003 | -0.5270 | No | ||
| 45 | CXCL12 | 580546 4150750 4570068 | 11890 | -0.003 | -0.5332 | No | ||
| 46 | CRK | 1230162 4780128 | 11959 | -0.004 | -0.5367 | No | ||
| 47 | AUH | 5570152 | 11975 | -0.004 | -0.5374 | No | ||
| 48 | NRK | 4590609 | 12243 | -0.005 | -0.5516 | No | ||
| 49 | KCNJ3 | 50184 | 12453 | -0.006 | -0.5626 | No | ||
| 50 | TBPL1 | 3610332 | 12665 | -0.007 | -0.5737 | No | ||
| 51 | WTAP | 4010176 | 12977 | -0.008 | -0.5901 | No | ||
| 52 | UHRF2 | 60050 840280 1340746 5080091 | 13068 | -0.009 | -0.5946 | No | ||
| 53 | SLITRK2 | 2640020 | 13159 | -0.009 | -0.5990 | No | ||
| 54 | STX12 | 610451 | 13242 | -0.010 | -0.6030 | No | ||
| 55 | FGD4 | 520168 870411 2640253 6550338 6650364 | 13313 | -0.010 | -0.6063 | No | ||
| 56 | HIPK1 | 110193 | 13664 | -0.013 | -0.6246 | No | ||
| 57 | RSBN1 | 7000487 | 13919 | -0.016 | -0.6377 | No | ||
| 58 | LRRC4 | 3850041 | 13929 | -0.016 | -0.6374 | No | ||
| 59 | PPARGC1A | 4670040 | 13963 | -0.016 | -0.6385 | No | ||
| 60 | POU4F2 | 2120195 2570022 | 14328 | -0.021 | -0.6572 | No | ||
| 61 | MAP4K4 | 2360059 | 14333 | -0.021 | -0.6564 | No | ||
| 62 | KPNA3 | 7040088 | 14468 | -0.023 | -0.6626 | No | ||
| 63 | NR4A3 | 2900021 5860095 5910039 | 14482 | -0.024 | -0.6622 | No | ||
| 64 | CDKN1B | 3800025 6450044 | 14515 | -0.024 | -0.6629 | No | ||
| 65 | TGFA | 1570332 | 14731 | -0.029 | -0.6732 | No | ||
| 66 | HOOK3 | 1190242 2470059 | 14853 | -0.033 | -0.6782 | No | ||
| 67 | CLASP1 | 6860279 | 15001 | -0.037 | -0.6844 | No | ||
| 68 | CHCHD4 | 2680075 | 15395 | -0.054 | -0.7032 | No | ||
| 69 | CREB1 | 1500717 2230358 3610600 6550601 | 15564 | -0.065 | -0.7094 | No | ||
| 70 | MRC1 | 730097 | 15805 | -0.081 | -0.7187 | No | ||
| 71 | PKP4 | 3710400 5890097 | 15807 | -0.081 | -0.7150 | No | ||
| 72 | TOP2B | 6980309 | 16219 | -0.116 | -0.7320 | Yes | ||
| 73 | HS2ST1 | 3170088 | 16325 | -0.127 | -0.7318 | Yes | ||
| 74 | NFAT5 | 2510411 5890195 6550152 | 16428 | -0.140 | -0.7310 | Yes | ||
| 75 | TRIM33 | 580619 2230280 3990433 6200747 | 16523 | -0.154 | -0.7291 | Yes | ||
| 76 | RPS6KA3 | 1980707 | 16549 | -0.158 | -0.7233 | Yes | ||
| 77 | ASF1A | 7050020 | 16589 | -0.163 | -0.7180 | Yes | ||
| 78 | DCUN1D1 | 2360332 | 16631 | -0.171 | -0.7124 | Yes | ||
| 79 | REV3L | 1090717 | 16742 | -0.185 | -0.7099 | Yes | ||
| 80 | ULK2 | 50041 5050176 | 16846 | -0.206 | -0.7062 | Yes | ||
| 81 | UTX | 2900017 6770452 | 16924 | -0.218 | -0.7004 | Yes | ||
| 82 | TMPO | 4050494 | 17165 | -0.266 | -0.7013 | Yes | ||
| 83 | NFIX | 2450152 | 17251 | -0.289 | -0.6928 | Yes | ||
| 84 | ATP11B | 4230100 | 17266 | -0.294 | -0.6802 | Yes | ||
| 85 | ENTPD5 | 4280070 | 17316 | -0.308 | -0.6689 | Yes | ||
| 86 | CSNK1G3 | 110450 | 17530 | -0.364 | -0.6638 | Yes | ||
| 87 | PJA2 | 430537 | 17555 | -0.371 | -0.6483 | Yes | ||
| 88 | CEPT1 | 1940047 | 17559 | -0.372 | -0.6316 | Yes | ||
| 89 | PDCD10 | 4760500 | 17634 | -0.404 | -0.6172 | Yes | ||
| 90 | DDX3X | 2190020 | 17682 | -0.421 | -0.6007 | Yes | ||
| 91 | ZDHHC21 | 2370338 | 17765 | -0.449 | -0.5847 | Yes | ||
| 92 | ARFIP1 | 3130601 | 17790 | -0.459 | -0.5651 | Yes | ||
| 93 | PITPNA | 2760332 | 17831 | -0.477 | -0.5456 | Yes | ||
| 94 | RANBP9 | 4670685 | 17870 | -0.489 | -0.5255 | Yes | ||
| 95 | CUL1 | 1990632 | 17893 | -0.503 | -0.5038 | Yes | ||
| 96 | SMARCAD1 | 5390619 | 17926 | -0.518 | -0.4820 | Yes | ||
| 97 | RAB1A | 2370671 | 17979 | -0.543 | -0.4602 | Yes | ||
| 98 | NDEL1 | 2370465 | 17995 | -0.554 | -0.4358 | Yes | ||
| 99 | ZHX1 | 2100110 2120286 | 18084 | -0.606 | -0.4131 | Yes | ||
| 100 | KPNA1 | 5270324 | 18099 | -0.612 | -0.3861 | Yes | ||
| 101 | UBL3 | 6450458 | 18155 | -0.645 | -0.3597 | Yes | ||
| 102 | DHX36 | 2470465 | 18180 | -0.659 | -0.3311 | Yes | ||
| 103 | FEM1B | 1230110 | 18181 | -0.659 | -0.3012 | Yes | ||
| 104 | SUMO1 | 1990364 2760338 6380195 | 18227 | -0.696 | -0.2720 | Yes | ||
| 105 | MAPRE1 | 3290037 | 18295 | -0.767 | -0.2408 | Yes | ||
| 106 | SATB1 | 5670154 | 18438 | -0.951 | -0.2053 | Yes | ||
| 107 | SFRS1 | 2360440 | 18508 | -1.134 | -0.1575 | Yes | ||
| 108 | TNRC6A | 1240537 6130037 6200010 | 18587 | -1.794 | -0.0803 | Yes | ||
| 109 | CCND1 | 460524 770309 3120576 6980398 | 18589 | -1.801 | 0.0015 | Yes |