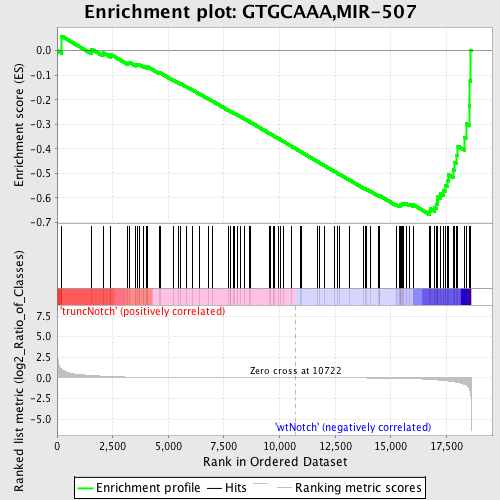
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | GTGCAAA,MIR-507 |
| Enrichment Score (ES) | -0.6671971 |
| Normalized Enrichment Score (NES) | -1.6489655 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.032540556 |
| FWER p-Value | 0.243 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HRB | 1660397 6110398 | 206 | 1.037 | 0.0590 | No | ||
| 2 | MAP3K3 | 610685 | 1546 | 0.286 | 0.0062 | No | ||
| 3 | TRAF7 | 1190341 | 2067 | 0.207 | -0.0079 | No | ||
| 4 | EMILIN2 | 70253 | 2417 | 0.162 | -0.0158 | No | ||
| 5 | RAPH1 | 6760411 | 3180 | 0.096 | -0.0504 | No | ||
| 6 | TMEM16D | 2650324 | 3255 | 0.090 | -0.0483 | No | ||
| 7 | BARHL1 | 4210113 | 3523 | 0.076 | -0.0576 | No | ||
| 8 | MIP | 6520333 | 3606 | 0.072 | -0.0571 | No | ||
| 9 | FBXW7 | 4210338 7050280 | 3694 | 0.069 | -0.0572 | No | ||
| 10 | CNIH | 2190484 5080609 | 3901 | 0.060 | -0.0643 | No | ||
| 11 | NELL2 | 2340725 4280301 5420008 | 4028 | 0.057 | -0.0672 | No | ||
| 12 | PPP1CB | 3130411 3780270 4150180 | 4083 | 0.055 | -0.0664 | No | ||
| 13 | RAB21 | 5570070 | 4606 | 0.041 | -0.0918 | No | ||
| 14 | CENTG2 | 4210053 | 4635 | 0.040 | -0.0906 | No | ||
| 15 | BACH2 | 6760131 | 5241 | 0.030 | -0.1212 | No | ||
| 16 | OLFML2B | 4060056 | 5440 | 0.027 | -0.1300 | No | ||
| 17 | STAT5B | 6200026 | 5565 | 0.026 | -0.1350 | No | ||
| 18 | DLL1 | 1770377 | 5809 | 0.023 | -0.1466 | No | ||
| 19 | NEUROD1 | 3060619 | 6106 | 0.020 | -0.1612 | No | ||
| 20 | SEMA6D | 4050324 5860138 6350307 | 6411 | 0.017 | -0.1764 | No | ||
| 21 | GDF6 | 1340048 | 6783 | 0.015 | -0.1955 | No | ||
| 22 | BCL11A | 6860369 | 6996 | 0.013 | -0.2060 | No | ||
| 23 | WDR26 | 1090138 | 7720 | 0.010 | -0.2444 | No | ||
| 24 | PRDM1 | 3170347 3520301 | 7781 | 0.009 | -0.2470 | No | ||
| 25 | MAB21L1 | 1450634 1580092 | 7800 | 0.009 | -0.2474 | No | ||
| 26 | PAX6 | 1190025 | 7910 | 0.009 | -0.2526 | No | ||
| 27 | NKX2-2 | 4150731 | 7976 | 0.009 | -0.2556 | No | ||
| 28 | MFN1 | 5080546 6200292 | 8096 | 0.008 | -0.2614 | No | ||
| 29 | MEIS1 | 1400575 | 8236 | 0.008 | -0.2684 | No | ||
| 30 | PCDH9 | 2100136 | 8403 | 0.007 | -0.2769 | No | ||
| 31 | RBMS3 | 6110722 | 8664 | 0.006 | -0.2905 | No | ||
| 32 | PRKCE | 5700053 | 8671 | 0.006 | -0.2905 | No | ||
| 33 | LHX8 | 5700347 | 9542 | 0.003 | -0.3372 | No | ||
| 34 | KCND2 | 1170128 2100112 | 9611 | 0.003 | -0.3406 | No | ||
| 35 | FMN2 | 450463 | 9728 | 0.003 | -0.3467 | No | ||
| 36 | PTPN2 | 130315 | 9772 | 0.003 | -0.3488 | No | ||
| 37 | ESRRG | 4010181 | 9935 | 0.002 | -0.3574 | No | ||
| 38 | ZIC1 | 670113 | 10040 | 0.002 | -0.3629 | No | ||
| 39 | RPS6KB1 | 1450427 5080110 6200563 | 10168 | 0.002 | -0.3697 | No | ||
| 40 | SLC24A4 | 2100487 | 10526 | 0.001 | -0.3889 | No | ||
| 41 | DAZL | 4050082 | 10933 | -0.001 | -0.4108 | No | ||
| 42 | SP3 | 3840338 | 10972 | -0.001 | -0.4128 | No | ||
| 43 | NDFIP1 | 1980402 | 11693 | -0.003 | -0.4514 | No | ||
| 44 | MAMDC1 | 3710129 4480075 | 11789 | -0.003 | -0.4563 | No | ||
| 45 | GRIK2 | 4610164 | 11999 | -0.004 | -0.4674 | No | ||
| 46 | KCNJ3 | 50184 | 12453 | -0.006 | -0.4914 | No | ||
| 47 | BMP4 | 380113 | 12597 | -0.006 | -0.4987 | No | ||
| 48 | SCN3A | 4070309 | 12707 | -0.007 | -0.5041 | No | ||
| 49 | ATP2B2 | 3780397 | 13136 | -0.009 | -0.5266 | No | ||
| 50 | TIAL1 | 4150048 6510605 | 13782 | -0.014 | -0.5605 | No | ||
| 51 | RGS17 | 1400603 | 13849 | -0.015 | -0.5630 | No | ||
| 52 | RSBN1 | 7000487 | 13919 | -0.016 | -0.5657 | No | ||
| 53 | SORCS1 | 60411 5890373 | 14083 | -0.018 | -0.5733 | No | ||
| 54 | THRAP2 | 4920600 | 14429 | -0.023 | -0.5904 | No | ||
| 55 | USP47 | 2640497 | 14474 | -0.024 | -0.5912 | No | ||
| 56 | NR4A3 | 2900021 5860095 5910039 | 14482 | -0.024 | -0.5899 | No | ||
| 57 | CHD9 | 430037 2570129 5420398 | 15256 | -0.048 | -0.6284 | No | ||
| 58 | BICD2 | 1090411 | 15367 | -0.053 | -0.6308 | No | ||
| 59 | BRD4 | 580471 610255 730537 1090132 1410204 1990242 2680088 3290019 4280706 | 15442 | -0.056 | -0.6310 | No | ||
| 60 | RAB22A | 110500 3830707 | 15447 | -0.057 | -0.6273 | No | ||
| 61 | THRA | 6770341 110164 1990600 | 15479 | -0.059 | -0.6250 | No | ||
| 62 | PCF11 | 1240180 | 15502 | -0.061 | -0.6221 | No | ||
| 63 | MDM2 | 3450053 5080138 | 15571 | -0.065 | -0.6214 | No | ||
| 64 | DYRK1A | 3190181 | 15694 | -0.073 | -0.6230 | No | ||
| 65 | STAG2 | 4540132 | 15837 | -0.084 | -0.6250 | No | ||
| 66 | PARP16 | 1690390 | 16016 | -0.097 | -0.6281 | No | ||
| 67 | REV3L | 1090717 | 16742 | -0.185 | -0.6546 | Yes | ||
| 68 | DNAJC13 | 4730091 | 16770 | -0.190 | -0.6432 | Yes | ||
| 69 | EDARADD | 840180 2360435 4560014 | 16970 | -0.226 | -0.6387 | Yes | ||
| 70 | DHX30 | 2480494 | 17059 | -0.244 | -0.6269 | Yes | ||
| 71 | SLC4A7 | 2370056 | 17088 | -0.250 | -0.6115 | Yes | ||
| 72 | LEF1 | 2470082 7100288 | 17114 | -0.255 | -0.5956 | Yes | ||
| 73 | XPO4 | 3840551 | 17226 | -0.281 | -0.5825 | Yes | ||
| 74 | MARK3 | 2370450 | 17373 | -0.324 | -0.5685 | Yes | ||
| 75 | NCOA1 | 3610438 | 17461 | -0.346 | -0.5498 | Yes | ||
| 76 | PJA2 | 430537 | 17555 | -0.371 | -0.5297 | Yes | ||
| 77 | GTF2H1 | 2570746 2650148 | 17587 | -0.381 | -0.5055 | Yes | ||
| 78 | CXXC6 | 2810168 | 17810 | -0.469 | -0.4858 | Yes | ||
| 79 | ATP1B1 | 3130594 | 17850 | -0.483 | -0.4552 | Yes | ||
| 80 | ZFX | 5900400 | 17969 | -0.540 | -0.4250 | Yes | ||
| 81 | RUNX1 | 3840711 | 18014 | -0.563 | -0.3893 | Yes | ||
| 82 | DAB2IP | 3170064 6200441 | 18308 | -0.779 | -0.3524 | Yes | ||
| 83 | MBNL1 | 2640762 7100048 | 18381 | -0.865 | -0.2978 | Yes | ||
| 84 | EIF4G2 | 3800575 6860184 | 18522 | -1.204 | -0.2240 | Yes | ||
| 85 | CDKN1C | 6520577 | 18557 | -1.527 | -0.1225 | Yes | ||
| 86 | CNOT7 | 2450338 5720397 | 18593 | -1.857 | 0.0012 | Yes |

