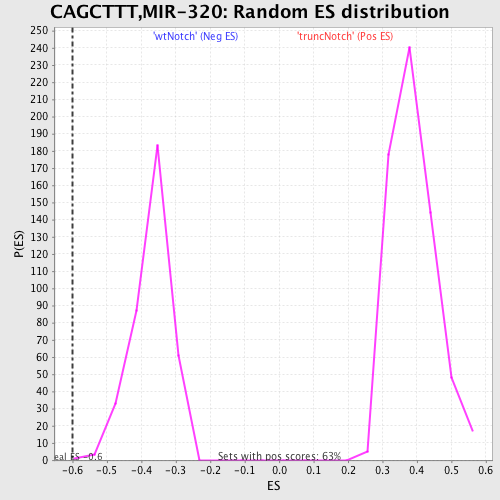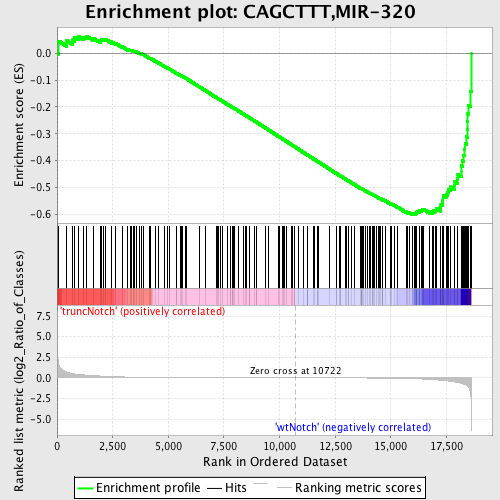
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | CAGCTTT,MIR-320 |
| Enrichment Score (ES) | -0.6013576 |
| Normalized Enrichment Score (NES) | -1.6285272 |
| Nominal p-value | 0.0027173914 |
| FDR q-value | 0.038921755 |
| FWER p-Value | 0.319 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NCDN | 50040 1980739 3520603 | 77 | 1.599 | 0.0455 | No | ||
| 2 | PPM1B | 380180 450154 3610215 | 435 | 0.684 | 0.0474 | No | ||
| 3 | RHOG | 6760575 | 684 | 0.516 | 0.0499 | No | ||
| 4 | PLK3 | 2640592 | 782 | 0.475 | 0.0594 | No | ||
| 5 | PLK2 | 6450152 | 956 | 0.416 | 0.0630 | No | ||
| 6 | DHDDS | 940148 6650184 | 1197 | 0.355 | 0.0610 | No | ||
| 7 | SLC10A3 | 2470563 | 1325 | 0.330 | 0.0643 | No | ||
| 8 | BHLHB2 | 7040603 | 1637 | 0.269 | 0.0558 | No | ||
| 9 | MRPS25 | 610441 870128 1410368 | 1942 | 0.223 | 0.0463 | No | ||
| 10 | ARMCX2 | 2320167 | 1972 | 0.219 | 0.0515 | No | ||
| 11 | TRAF7 | 1190341 | 2067 | 0.207 | 0.0528 | No | ||
| 12 | FKBP1A | 2450368 | 2169 | 0.193 | 0.0534 | No | ||
| 13 | SON | 3120148 5860239 7040403 | 2451 | 0.158 | 0.0430 | No | ||
| 14 | IGF1R | 3360494 | 2645 | 0.138 | 0.0369 | No | ||
| 15 | DAB2 | 60309 | 2928 | 0.112 | 0.0250 | No | ||
| 16 | TNFRSF21 | 6380100 | 3175 | 0.096 | 0.0147 | No | ||
| 17 | PHF1 | 4010577 6420053 | 3283 | 0.089 | 0.0117 | No | ||
| 18 | KCNH7 | 7510068 | 3329 | 0.086 | 0.0119 | No | ||
| 19 | CDK6 | 4920253 | 3448 | 0.079 | 0.0079 | No | ||
| 20 | PCDHA11 | 1510253 | 3488 | 0.078 | 0.0082 | No | ||
| 21 | NR4A2 | 60273 | 3577 | 0.073 | 0.0057 | No | ||
| 22 | ATXN1 | 5550156 | 3707 | 0.068 | 0.0009 | No | ||
| 23 | GSPT2 | 6590674 | 3790 | 0.065 | -0.0016 | No | ||
| 24 | MXI1 | 5050064 5130484 | 3870 | 0.061 | -0.0039 | No | ||
| 25 | UNC5A | 2120528 3060324 | 4149 | 0.053 | -0.0174 | No | ||
| 26 | LPHN1 | 1400113 3130301 | 4202 | 0.051 | -0.0186 | No | ||
| 27 | A2BP1 | 2370390 4590593 5550014 | 4412 | 0.046 | -0.0285 | No | ||
| 28 | PCDHA5 | 2970347 | 4570 | 0.042 | -0.0357 | No | ||
| 29 | PCSK7 | 6370450 | 4822 | 0.037 | -0.0482 | No | ||
| 30 | HIVEP2 | 6520538 | 4959 | 0.035 | -0.0545 | No | ||
| 31 | HECTD2 | 2680014 3060689 | 5070 | 0.033 | -0.0594 | No | ||
| 32 | ABCA12 | 4540156 | 5364 | 0.028 | -0.0744 | No | ||
| 33 | CDH2 | 520435 2450451 2760025 6650477 | 5382 | 0.028 | -0.0745 | No | ||
| 34 | PCDHA6 | 780333 2370280 | 5532 | 0.026 | -0.0818 | No | ||
| 35 | ARID5B | 110403 | 5568 | 0.026 | -0.0829 | No | ||
| 36 | CREB5 | 2320368 | 5656 | 0.024 | -0.0868 | No | ||
| 37 | SMCR8 | 730653 | 5790 | 0.023 | -0.0933 | No | ||
| 38 | PPP2R5B | 2900441 | 5807 | 0.023 | -0.0935 | No | ||
| 39 | SEMA6D | 4050324 5860138 6350307 | 6411 | 0.017 | -0.1256 | No | ||
| 40 | DBN1 | 7100537 | 6650 | 0.015 | -0.1381 | No | ||
| 41 | RAI2 | 6770333 | 7157 | 0.012 | -0.1651 | No | ||
| 42 | BMPR1A | 2190193 4060603 | 7209 | 0.012 | -0.1675 | No | ||
| 43 | CALD1 | 1770129 1940397 | 7234 | 0.012 | -0.1684 | No | ||
| 44 | FLRT3 | 70685 2760497 6040519 | 7341 | 0.011 | -0.1738 | No | ||
| 45 | PAK7 | 3830164 | 7423 | 0.011 | -0.1779 | No | ||
| 46 | NAP1L5 | 360576 | 7677 | 0.010 | -0.1913 | No | ||
| 47 | RAB18 | 3120575 4590577 | 7771 | 0.009 | -0.1960 | No | ||
| 48 | NPAS2 | 770670 | 7804 | 0.009 | -0.1975 | No | ||
| 49 | TRIM41 | 4070019 6110670 | 7868 | 0.009 | -0.2006 | No | ||
| 50 | ENAH | 1690292 5700300 | 7886 | 0.009 | -0.2013 | No | ||
| 51 | CALN1 | 3610722 | 7913 | 0.009 | -0.2024 | No | ||
| 52 | RKHD3 | 6180471 | 7952 | 0.009 | -0.2042 | No | ||
| 53 | PCDHA12 | 2810215 | 8164 | 0.008 | -0.2154 | No | ||
| 54 | IPO7 | 2190746 | 8359 | 0.007 | -0.2257 | No | ||
| 55 | PLXNC1 | 5860315 | 8449 | 0.007 | -0.2303 | No | ||
| 56 | PHOX2B | 5270075 | 8527 | 0.006 | -0.2343 | No | ||
| 57 | POGZ | 870348 7040286 | 8649 | 0.006 | -0.2406 | No | ||
| 58 | RBMS3 | 6110722 | 8664 | 0.006 | -0.2412 | No | ||
| 59 | EIF3S1 | 6130368 6770044 | 8891 | 0.005 | -0.2533 | No | ||
| 60 | RAP1A | 1090025 | 8964 | 0.005 | -0.2571 | No | ||
| 61 | PCDHA10 | 3830725 | 8970 | 0.005 | -0.2572 | No | ||
| 62 | CHES1 | 4570121 5700162 | 9351 | 0.004 | -0.2777 | No | ||
| 63 | BANP | 940528 | 9521 | 0.003 | -0.2867 | No | ||
| 64 | ESRRG | 4010181 | 9935 | 0.002 | -0.3090 | No | ||
| 65 | BTBD3 | 1980041 2510577 3190053 4200746 | 9988 | 0.002 | -0.3118 | No | ||
| 66 | EYA4 | 5360286 | 10126 | 0.002 | -0.3192 | No | ||
| 67 | TAF5 | 3450288 5890193 6860435 | 10196 | 0.001 | -0.3229 | No | ||
| 68 | ADAM10 | 3780156 | 10223 | 0.001 | -0.3242 | No | ||
| 69 | TMEM47 | 1660086 | 10305 | 0.001 | -0.3286 | No | ||
| 70 | KCNS3 | 4150039 | 10524 | 0.001 | -0.3404 | No | ||
| 71 | ELL2 | 6420091 | 10575 | 0.000 | -0.3431 | No | ||
| 72 | WRNIP1 | 2320541 5890725 | 10647 | 0.000 | -0.3469 | No | ||
| 73 | EREG | 50519 4920129 | 10683 | 0.000 | -0.3488 | No | ||
| 74 | XPO1 | 540707 | 10862 | -0.000 | -0.3584 | No | ||
| 75 | CACNA1E | 840253 | 11094 | -0.001 | -0.3709 | No | ||
| 76 | MMP16 | 2680139 | 11236 | -0.001 | -0.3785 | No | ||
| 77 | KLF5 | 3840348 | 11247 | -0.001 | -0.3790 | No | ||
| 78 | CDH20 | 5420215 7100711 | 11271 | -0.002 | -0.3802 | No | ||
| 79 | ZIC3 | 380020 | 11531 | -0.002 | -0.3942 | No | ||
| 80 | AMBN | 4150170 | 11573 | -0.002 | -0.3964 | No | ||
| 81 | TDG | 2900358 2940079 | 11705 | -0.003 | -0.4034 | No | ||
| 82 | PLAG1 | 1450142 3870139 6520039 | 11708 | -0.003 | -0.4034 | No | ||
| 83 | RAD9A | 110300 1940632 3990390 6040014 | 11765 | -0.003 | -0.4063 | No | ||
| 84 | YES1 | 6100050 | 12234 | -0.005 | -0.4315 | No | ||
| 85 | SATB2 | 2470181 | 12540 | -0.006 | -0.4479 | No | ||
| 86 | PCDHA3 | 3840402 | 12568 | -0.006 | -0.4492 | No | ||
| 87 | CAB39 | 5720471 | 12673 | -0.007 | -0.4546 | No | ||
| 88 | ATRX | 2340039 3610132 4210373 | 12740 | -0.007 | -0.4580 | No | ||
| 89 | USP25 | 3520035 | 12750 | -0.007 | -0.4582 | No | ||
| 90 | TROVE2 | 4850458 | 12944 | -0.008 | -0.4684 | No | ||
| 91 | TSC1 | 1850672 | 13010 | -0.008 | -0.4717 | No | ||
| 92 | NEGR1 | 1940731 5130170 | 13107 | -0.009 | -0.4766 | No | ||
| 93 | ING5 | 3830440 5910239 6200471 6650576 | 13119 | -0.009 | -0.4769 | No | ||
| 94 | ARPC5 | 1740411 3450300 | 13252 | -0.010 | -0.4838 | No | ||
| 95 | DSCAM | 1780050 2450731 2810438 | 13347 | -0.011 | -0.4886 | No | ||
| 96 | PHF8 | 70722 | 13630 | -0.013 | -0.5035 | No | ||
| 97 | HIPK1 | 110193 | 13664 | -0.013 | -0.5048 | No | ||
| 98 | PCDHA1 | 940026 540484 | 13688 | -0.013 | -0.5057 | No | ||
| 99 | FOXQ1 | 4850088 | 13694 | -0.013 | -0.5055 | No | ||
| 100 | LRCH2 | 2900162 | 13738 | -0.014 | -0.5074 | No | ||
| 101 | AZIN1 | 3450070 | 13771 | -0.014 | -0.5087 | No | ||
| 102 | PBX3 | 1300424 3710577 6180575 | 13851 | -0.015 | -0.5126 | No | ||
| 103 | PPARGC1A | 4670040 | 13963 | -0.016 | -0.5181 | No | ||
| 104 | MSI2 | 1190066 5270039 | 14058 | -0.017 | -0.5227 | No | ||
| 105 | RHOBTB1 | 1410010 | 14088 | -0.018 | -0.5237 | No | ||
| 106 | PALM2 | 4120068 7100398 | 14167 | -0.019 | -0.5273 | No | ||
| 107 | SPRED2 | 510138 540195 | 14186 | -0.019 | -0.5277 | No | ||
| 108 | NONO | 7050014 | 14221 | -0.019 | -0.5290 | No | ||
| 109 | TMEM2 | 1980037 3290020 | 14265 | -0.020 | -0.5307 | No | ||
| 110 | DNER | 3610170 | 14342 | -0.021 | -0.5341 | No | ||
| 111 | MLLT3 | 510204 3850722 6200088 | 14464 | -0.023 | -0.5399 | No | ||
| 112 | QKI | 5220093 6130044 | 14486 | -0.024 | -0.5403 | No | ||
| 113 | ZNF3 | 1770193 2320047 2570273 | 14548 | -0.025 | -0.5429 | No | ||
| 114 | SLITRK3 | 5910041 | 14641 | -0.027 | -0.5470 | No | ||
| 115 | ICA1 | 3830059 | 14643 | -0.027 | -0.5462 | No | ||
| 116 | SDHC | 3140347 6100156 | 14647 | -0.027 | -0.5456 | No | ||
| 117 | YWHAE | 5310435 | 14779 | -0.030 | -0.5517 | No | ||
| 118 | DTNA | 1340600 1780731 2340278 2850132 | 14995 | -0.037 | -0.5622 | No | ||
| 119 | CLASP1 | 6860279 | 15001 | -0.037 | -0.5613 | No | ||
| 120 | AP3M1 | 5340433 | 15043 | -0.039 | -0.5623 | No | ||
| 121 | LUC7L2 | 1850402 6350088 | 15163 | -0.044 | -0.5674 | No | ||
| 122 | ULK1 | 6100315 | 15303 | -0.049 | -0.5734 | No | ||
| 123 | YTHDF3 | 4540632 4670717 7040609 | 15687 | -0.073 | -0.5919 | No | ||
| 124 | SMAP1 | 840056 2360523 | 15771 | -0.079 | -0.5940 | No | ||
| 125 | STAG2 | 4540132 | 15837 | -0.084 | -0.5949 | No | ||
| 126 | CPEB1 | 3190524 | 15956 | -0.092 | -0.5985 | Yes | ||
| 127 | GABPB2 | 6130450 7050735 | 15964 | -0.093 | -0.5959 | Yes | ||
| 128 | CALM3 | 3390288 | 16062 | -0.101 | -0.5981 | Yes | ||
| 129 | DAZAP1 | 5890563 | 16124 | -0.106 | -0.5981 | Yes | ||
| 130 | PCDHA7 | 1850019 | 16139 | -0.108 | -0.5955 | Yes | ||
| 131 | MAPK1 | 3190193 6200253 | 16153 | -0.109 | -0.5928 | Yes | ||
| 132 | YWHAQ | 6760524 | 16174 | -0.112 | -0.5904 | Yes | ||
| 133 | GRB2 | 6650398 | 16274 | -0.122 | -0.5920 | Yes | ||
| 134 | AIM1 | 2810142 | 16278 | -0.122 | -0.5883 | Yes | ||
| 135 | DAG1 | 460053 610341 | 16300 | -0.125 | -0.5856 | Yes | ||
| 136 | MAPK8IP3 | 1660112 2060427 | 16397 | -0.136 | -0.5866 | Yes | ||
| 137 | PURB | 5360138 | 16402 | -0.136 | -0.5826 | Yes | ||
| 138 | YWHAZ | 1230717 | 16461 | -0.145 | -0.5812 | Yes | ||
| 139 | RAD18 | 5670372 | 16747 | -0.186 | -0.5909 | Yes | ||
| 140 | PPP1R16B | 3520300 | 16886 | -0.212 | -0.5918 | Yes | ||
| 141 | SUHW3 | 50400 | 16911 | -0.216 | -0.5864 | Yes | ||
| 142 | MAT2A | 4070026 4730079 6020280 | 17019 | -0.235 | -0.5849 | Yes | ||
| 143 | DHX30 | 2480494 | 17059 | -0.244 | -0.5794 | Yes | ||
| 144 | SEP15 | 1780451 2060450 5890451 | 17231 | -0.283 | -0.5799 | Yes | ||
| 145 | TOMM70A | 780114 | 17248 | -0.287 | -0.5719 | Yes | ||
| 146 | MECP2 | 1940450 | 17253 | -0.290 | -0.5631 | Yes | ||
| 147 | TLK2 | 1170161 | 17328 | -0.312 | -0.5574 | Yes | ||
| 148 | PPM1A | 1170301 5900273 | 17340 | -0.317 | -0.5482 | Yes | ||
| 149 | POLE4 | 6860097 | 17345 | -0.317 | -0.5385 | Yes | ||
| 150 | HNRPC | 1570133 3610131 | 17366 | -0.323 | -0.5296 | Yes | ||
| 151 | SFRS7 | 2760408 | 17496 | -0.354 | -0.5256 | Yes | ||
| 152 | ARL8B | 3710368 | 17564 | -0.373 | -0.5177 | Yes | ||
| 153 | UBE2D3 | 3190452 | 17584 | -0.381 | -0.5069 | Yes | ||
| 154 | TPD52L2 | 6590040 | 17670 | -0.416 | -0.4985 | Yes | ||
| 155 | USP20 | 520128 | 17869 | -0.489 | -0.4941 | Yes | ||
| 156 | ATP6V1A | 6590242 | 17877 | -0.493 | -0.4792 | Yes | ||
| 157 | KLF13 | 4670563 | 17991 | -0.551 | -0.4682 | Yes | ||
| 158 | RUNX1 | 3840711 | 18014 | -0.563 | -0.4519 | Yes | ||
| 159 | OGT | 2360131 4610333 | 18167 | -0.653 | -0.4398 | Yes | ||
| 160 | ARPP-19 | 5700025 | 18169 | -0.654 | -0.4196 | Yes | ||
| 161 | GSPT1 | 5420050 | 18220 | -0.689 | -0.4009 | Yes | ||
| 162 | COPS2 | 3060142 | 18265 | -0.735 | -0.3805 | Yes | ||
| 163 | RASA1 | 1240315 | 18297 | -0.769 | -0.3583 | Yes | ||
| 164 | RAP2C | 1690132 | 18334 | -0.799 | -0.3354 | Yes | ||
| 165 | HOXB4 | 540131 | 18409 | -0.901 | -0.3115 | Yes | ||
| 166 | MCL1 | 1660672 | 18432 | -0.934 | -0.2836 | Yes | ||
| 167 | DHX15 | 870632 | 18439 | -0.952 | -0.2544 | Yes | ||
| 168 | CUTL1 | 1570113 1770091 1850273 2100286 5340605 | 18448 | -0.967 | -0.2248 | Yes | ||
| 169 | TFRC | 4050551 | 18490 | -1.089 | -0.1932 | Yes | ||
| 170 | CNOT7 | 2450338 5720397 | 18593 | -1.857 | -0.1411 | Yes | ||
| 171 | TPM3 | 670600 3170296 5670167 6650471 | 18614 | -4.583 | 0.0001 | Yes |