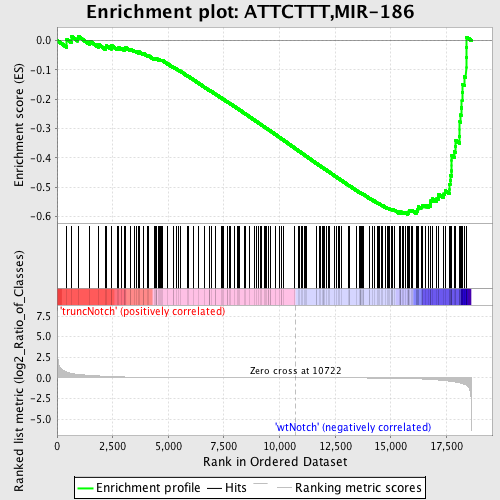
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | ATTCTTT,MIR-186 |
| Enrichment Score (ES) | -0.5925202 |
| Normalized Enrichment Score (NES) | -1.635767 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.038024243 |
| FWER p-Value | 0.3 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PPM1B | 380180 450154 3610215 | 435 | 0.684 | 0.0038 | No | ||
| 2 | LGTN | 450332 | 647 | 0.532 | 0.0136 | No | ||
| 3 | TARDBP | 870390 2350093 | 938 | 0.419 | 0.0147 | No | ||
| 4 | CXCL13 | 6290402 | 1472 | 0.300 | -0.0022 | No | ||
| 5 | EXT2 | 6520239 | 1869 | 0.235 | -0.0143 | No | ||
| 6 | RASA3 | 2060465 | 2192 | 0.190 | -0.0242 | No | ||
| 7 | HYOU1 | 3060465 | 2204 | 0.189 | -0.0172 | No | ||
| 8 | BCL9 | 7100112 | 2431 | 0.161 | -0.0230 | No | ||
| 9 | RBM16 | 6290286 6400181 | 2452 | 0.158 | -0.0178 | No | ||
| 10 | CPEB2 | 4760338 | 2715 | 0.130 | -0.0268 | No | ||
| 11 | FRMD4A | 2190176 | 2775 | 0.126 | -0.0250 | No | ||
| 12 | ZFAND3 | 2760504 | 2898 | 0.114 | -0.0270 | No | ||
| 13 | RABIF | 2370458 5910543 | 3017 | 0.105 | -0.0292 | No | ||
| 14 | DLG5 | 450215 | 3034 | 0.104 | -0.0259 | No | ||
| 15 | TMEM35 | 2640446 | 3086 | 0.101 | -0.0246 | No | ||
| 16 | DLC1 | 1090632 6450594 | 3281 | 0.089 | -0.0316 | No | ||
| 17 | BUB3 | 3170546 | 3320 | 0.087 | -0.0302 | No | ||
| 18 | PCDHA11 | 1510253 | 3488 | 0.078 | -0.0361 | No | ||
| 19 | NR4A2 | 60273 | 3577 | 0.073 | -0.0380 | No | ||
| 20 | EIF4E | 1580403 70133 6380215 | 3677 | 0.069 | -0.0406 | No | ||
| 21 | EIF3S10 | 1190079 3840176 | 3700 | 0.068 | -0.0390 | No | ||
| 22 | COL3A1 | 6420273 6620044 | 3873 | 0.061 | -0.0459 | No | ||
| 23 | GNAZ | 6130296 | 3892 | 0.061 | -0.0445 | No | ||
| 24 | PPP1CB | 3130411 3780270 4150180 | 4083 | 0.055 | -0.0526 | No | ||
| 25 | RBM9 | 2510092 6290088 | 4101 | 0.054 | -0.0513 | No | ||
| 26 | TRPS1 | 870100 | 4366 | 0.047 | -0.0638 | No | ||
| 27 | A2BP1 | 2370390 4590593 5550014 | 4412 | 0.046 | -0.0644 | No | ||
| 28 | RB1CC1 | 510494 7100072 | 4445 | 0.045 | -0.0643 | No | ||
| 29 | DOK6 | 4850673 | 4447 | 0.045 | -0.0626 | No | ||
| 30 | SPAST | 460603 4070471 4210132 4670338 | 4449 | 0.045 | -0.0608 | No | ||
| 31 | PCDHA5 | 2970347 | 4570 | 0.042 | -0.0657 | No | ||
| 32 | RAB21 | 5570070 | 4606 | 0.041 | -0.0659 | No | ||
| 33 | PPM1G | 610725 | 4659 | 0.040 | -0.0671 | No | ||
| 34 | JPH1 | 610739 | 4713 | 0.039 | -0.0685 | No | ||
| 35 | ZNF385 | 450176 4810358 | 4716 | 0.039 | -0.0670 | No | ||
| 36 | PAPPA | 4230463 | 4975 | 0.034 | -0.0796 | No | ||
| 37 | CNTNAP1 | 6650132 | 5235 | 0.030 | -0.0925 | No | ||
| 38 | BACH2 | 6760131 | 5241 | 0.030 | -0.0916 | No | ||
| 39 | CUGBP1 | 450292 510022 7050176 7050215 | 5365 | 0.028 | -0.0971 | No | ||
| 40 | HOXB8 | 3710450 | 5466 | 0.027 | -0.1015 | No | ||
| 41 | PCDHA6 | 780333 2370280 | 5532 | 0.026 | -0.1039 | No | ||
| 42 | SMPD3 | 520164 | 5877 | 0.022 | -0.1217 | No | ||
| 43 | PDGFC | 4200133 | 5889 | 0.022 | -0.1215 | No | ||
| 44 | NXPH1 | 3870546 | 6109 | 0.020 | -0.1326 | No | ||
| 45 | CALB1 | 460070 | 6374 | 0.018 | -0.1462 | No | ||
| 46 | MAP3K2 | 380270 | 6645 | 0.015 | -0.1602 | No | ||
| 47 | CNTNAP2 | 360170 4280546 | 6828 | 0.014 | -0.1695 | No | ||
| 48 | EPHB1 | 4610056 | 6830 | 0.014 | -0.1690 | No | ||
| 49 | SRPK1 | 450110 | 6847 | 0.014 | -0.1693 | No | ||
| 50 | TRPC1 | 3120189 4210537 | 6959 | 0.013 | -0.1748 | No | ||
| 51 | OSBPL8 | 50603 | 7136 | 0.012 | -0.1839 | No | ||
| 52 | WNK3 | 4060053 | 7139 | 0.012 | -0.1835 | No | ||
| 53 | SMAD6 | 870504 | 7368 | 0.011 | -0.1954 | No | ||
| 54 | PAK7 | 3830164 | 7423 | 0.011 | -0.1979 | No | ||
| 55 | LPHN2 | 4480010 | 7459 | 0.011 | -0.1994 | No | ||
| 56 | MCFD2 | 520576 5690017 | 7654 | 0.010 | -0.2095 | No | ||
| 57 | CPEB3 | 3940164 | 7663 | 0.010 | -0.2095 | No | ||
| 58 | IL2 | 1770725 | 7742 | 0.009 | -0.2134 | No | ||
| 59 | PRDM1 | 3170347 3520301 | 7781 | 0.009 | -0.2151 | No | ||
| 60 | RKHD3 | 6180471 | 7952 | 0.009 | -0.2240 | No | ||
| 61 | TPR | 1990066 6550390 | 8106 | 0.008 | -0.2319 | No | ||
| 62 | PCDHA12 | 2810215 | 8164 | 0.008 | -0.2347 | No | ||
| 63 | IGSF11 | 60471 2350288 | 8184 | 0.008 | -0.2354 | No | ||
| 64 | CXXC5 | 840010 | 8418 | 0.007 | -0.2478 | No | ||
| 65 | JAG1 | 3440390 | 8486 | 0.007 | -0.2512 | No | ||
| 66 | WDFY3 | 3610041 4560333 4570273 | 8653 | 0.006 | -0.2600 | No | ||
| 67 | NR3C2 | 2480324 6400093 | 8892 | 0.005 | -0.2727 | No | ||
| 68 | PCDHA10 | 3830725 | 8970 | 0.005 | -0.2766 | No | ||
| 69 | LMBR1 | 4570072 | 9040 | 0.005 | -0.2802 | No | ||
| 70 | UBQLN2 | 430397 1170594 4070121 | 9126 | 0.005 | -0.2846 | No | ||
| 71 | GRIN3A | 2370592 | 9203 | 0.004 | -0.2886 | No | ||
| 72 | DNM3 | 730711 | 9320 | 0.004 | -0.2947 | No | ||
| 73 | JPH3 | 780397 | 9344 | 0.004 | -0.2958 | No | ||
| 74 | ZCCHC14 | 5900288 | 9392 | 0.004 | -0.2982 | No | ||
| 75 | CLDN1 | 5670746 | 9489 | 0.003 | -0.3033 | No | ||
| 76 | CACNB3 | 2480035 4540184 | 9501 | 0.003 | -0.3037 | No | ||
| 77 | CD200R1 | 630593 | 9610 | 0.003 | -0.3095 | No | ||
| 78 | KCND2 | 1170128 2100112 | 9611 | 0.003 | -0.3094 | No | ||
| 79 | LRRN1 | 3290154 | 9805 | 0.003 | -0.3197 | No | ||
| 80 | BTBD3 | 1980041 2510577 3190053 4200746 | 9988 | 0.002 | -0.3295 | No | ||
| 81 | SP4 | 3850176 | 10068 | 0.002 | -0.3337 | No | ||
| 82 | ZFHX4 | 6110167 | 10098 | 0.002 | -0.3352 | No | ||
| 83 | RPS6KB1 | 1450427 5080110 6200563 | 10168 | 0.002 | -0.3389 | No | ||
| 84 | ZFP36L1 | 2510138 4120048 | 10657 | 0.000 | -0.3654 | No | ||
| 85 | PNN | 6650504 | 10833 | -0.000 | -0.3749 | No | ||
| 86 | PSPH | 4050020 | 10900 | -0.000 | -0.3785 | No | ||
| 87 | SP3 | 3840338 | 10972 | -0.001 | -0.3823 | No | ||
| 88 | MYT1L | 5670369 6660403 | 11048 | -0.001 | -0.3863 | No | ||
| 89 | TLOC1 | 4050309 | 11140 | -0.001 | -0.3912 | No | ||
| 90 | FOXD1 | 6860053 | 11158 | -0.001 | -0.3921 | No | ||
| 91 | KCNMA1 | 1450402 | 11220 | -0.001 | -0.3953 | No | ||
| 92 | SALL1 | 5420020 7050195 | 11659 | -0.003 | -0.4190 | No | ||
| 93 | MYT1 | 6770524 | 11671 | -0.003 | -0.4195 | No | ||
| 94 | ZCCHC11 | 3390731 5360039 | 11774 | -0.003 | -0.4249 | No | ||
| 95 | EFEMP1 | 5860324 | 11829 | -0.003 | -0.4277 | No | ||
| 96 | YY1 | 6130451 | 11945 | -0.004 | -0.4338 | No | ||
| 97 | CRK | 1230162 4780128 | 11959 | -0.004 | -0.4344 | No | ||
| 98 | GABRA4 | 6290465 | 11966 | -0.004 | -0.4345 | No | ||
| 99 | MITF | 380056 | 12033 | -0.004 | -0.4380 | No | ||
| 100 | INSM1 | 3450671 | 12090 | -0.004 | -0.4408 | No | ||
| 101 | SNAP91 | 3170056 5690520 | 12200 | -0.005 | -0.4466 | No | ||
| 102 | PLS3 | 3290082 | 12242 | -0.005 | -0.4486 | No | ||
| 103 | UBE2B | 4780497 | 12466 | -0.006 | -0.4605 | No | ||
| 104 | PCDHA3 | 3840402 | 12568 | -0.006 | -0.4657 | No | ||
| 105 | SOX11 | 610279 | 12649 | -0.007 | -0.4698 | No | ||
| 106 | HNMT | 4560022 | 12689 | -0.007 | -0.4716 | No | ||
| 107 | LIN28 | 6590672 | 12789 | -0.007 | -0.4767 | No | ||
| 108 | NEGR1 | 1940731 5130170 | 13107 | -0.009 | -0.4936 | No | ||
| 109 | AMOT | 1940121 2940647 6590093 | 13125 | -0.009 | -0.4941 | No | ||
| 110 | BIRC4 | 1400129 6960128 | 13150 | -0.009 | -0.4950 | No | ||
| 111 | DPP9 | 6660441 | 13439 | -0.011 | -0.5102 | No | ||
| 112 | PSMD11 | 2340538 6510053 | 13573 | -0.012 | -0.5170 | No | ||
| 113 | BCAR3 | 1660717 | 13614 | -0.013 | -0.5186 | No | ||
| 114 | DDX3Y | 1580278 4200519 | 13631 | -0.013 | -0.5190 | No | ||
| 115 | CAPZA1 | 3140390 | 13647 | -0.013 | -0.5193 | No | ||
| 116 | PCDHA1 | 940026 540484 | 13688 | -0.013 | -0.5209 | No | ||
| 117 | LRCH2 | 2900162 | 13738 | -0.014 | -0.5230 | No | ||
| 118 | OTX1 | 2510215 | 13757 | -0.014 | -0.5235 | No | ||
| 119 | WNT5A | 840685 3120152 | 13766 | -0.014 | -0.5233 | No | ||
| 120 | SENP6 | 1340592 4730142 | 14051 | -0.017 | -0.5381 | No | ||
| 121 | ITGA6 | 3830129 | 14056 | -0.017 | -0.5376 | No | ||
| 122 | CCNT2 | 2100390 4150563 | 14196 | -0.019 | -0.5444 | No | ||
| 123 | CSNK2A1 | 1580577 | 14243 | -0.020 | -0.5461 | No | ||
| 124 | NCOA2 | 940347 2850725 | 14393 | -0.022 | -0.5533 | No | ||
| 125 | HOXA9 | 610494 4730040 5130601 | 14442 | -0.023 | -0.5550 | No | ||
| 126 | QKI | 5220093 6130044 | 14486 | -0.024 | -0.5564 | No | ||
| 127 | EPN2 | 1340451 6550239 6860020 6860403 | 14601 | -0.026 | -0.5615 | No | ||
| 128 | SEPT7 | 2760685 | 14644 | -0.027 | -0.5627 | No | ||
| 129 | CDC5L | 3130446 3390025 6400204 | 14776 | -0.030 | -0.5686 | No | ||
| 130 | HOOK3 | 1190242 2470059 | 14853 | -0.033 | -0.5714 | No | ||
| 131 | ZZZ3 | 780133 2320239 2640452 5080706 6550341 | 14878 | -0.033 | -0.5714 | No | ||
| 132 | RAB6A | 3840463 4780731 | 14953 | -0.035 | -0.5740 | No | ||
| 133 | CAST | 3850082 5420671 6200156 | 15015 | -0.038 | -0.5758 | No | ||
| 134 | NR5A2 | 1690180 6900050 | 15036 | -0.039 | -0.5753 | No | ||
| 135 | NME7 | 60170 1230551 2850333 4200102 4560411 5090647 | 15075 | -0.040 | -0.5758 | No | ||
| 136 | FNDC3A | 2690097 3290044 | 15085 | -0.040 | -0.5746 | No | ||
| 137 | ACSL4 | 510020 1240041 3190050 4540500 5570463 | 15179 | -0.044 | -0.5779 | No | ||
| 138 | EFCBP1 | 5860450 | 15396 | -0.054 | -0.5875 | No | ||
| 139 | MOSPD1 | 2030170 5270156 6900142 | 15423 | -0.056 | -0.5866 | No | ||
| 140 | PTGES3 | 2190440 2680524 | 15425 | -0.056 | -0.5845 | No | ||
| 141 | GPR85 | 5130750 5910020 | 15438 | -0.056 | -0.5829 | No | ||
| 142 | PPP1CC | 6380300 2510647 | 15544 | -0.063 | -0.5860 | No | ||
| 143 | SYAP1 | 1410500 | 15574 | -0.065 | -0.5850 | No | ||
| 144 | SUCLA2 | 730632 1170292 | 15647 | -0.070 | -0.5861 | No | ||
| 145 | SLC25A3 | 670112 | 15766 | -0.079 | -0.5894 | Yes | ||
| 146 | UBE3A | 1240152 2690438 5860609 | 15786 | -0.080 | -0.5872 | Yes | ||
| 147 | NARG1 | 5910563 6350095 | 15804 | -0.081 | -0.5849 | Yes | ||
| 148 | PRPF39 | 1090088 | 15815 | -0.082 | -0.5822 | Yes | ||
| 149 | STAG2 | 4540132 | 15837 | -0.084 | -0.5799 | Yes | ||
| 150 | PSME3 | 2810537 | 15913 | -0.089 | -0.5804 | Yes | ||
| 151 | GABPB2 | 6130450 7050735 | 15964 | -0.093 | -0.5794 | Yes | ||
| 152 | PCDHA7 | 1850019 | 16139 | -0.108 | -0.5846 | Yes | ||
| 153 | MAPK1 | 3190193 6200253 | 16153 | -0.109 | -0.5809 | Yes | ||
| 154 | MRPL3 | 1170736 5270025 | 16209 | -0.115 | -0.5792 | Yes | ||
| 155 | FBXO33 | 1510504 | 16211 | -0.115 | -0.5747 | Yes | ||
| 156 | TCF20 | 1190338 1240520 4070594 4810440 | 16222 | -0.117 | -0.5705 | Yes | ||
| 157 | ENC1 | 4810348 | 16223 | -0.117 | -0.5659 | Yes | ||
| 158 | BTF3 | 6940279 | 16358 | -0.131 | -0.5679 | Yes | ||
| 159 | PURB | 5360138 | 16402 | -0.136 | -0.5648 | Yes | ||
| 160 | NFAT5 | 2510411 5890195 6550152 | 16428 | -0.140 | -0.5605 | Yes | ||
| 161 | RPS6KA3 | 1980707 | 16549 | -0.158 | -0.5607 | Yes | ||
| 162 | P2RX7 | 6020048 6380053 6510121 | 16708 | -0.182 | -0.5620 | Yes | ||
| 163 | LRBA | 3450402 | 16772 | -0.191 | -0.5578 | Yes | ||
| 164 | TGFBR2 | 1780711 1980537 6550398 | 16790 | -0.195 | -0.5509 | Yes | ||
| 165 | HNRPU | 6520736 | 16801 | -0.196 | -0.5436 | Yes | ||
| 166 | CTSB | 1400524 2360064 | 16878 | -0.211 | -0.5393 | Yes | ||
| 167 | SLC23A2 | 6660722 | 17066 | -0.245 | -0.5396 | Yes | ||
| 168 | MAPKAP1 | 1660161 2450575 | 17130 | -0.259 | -0.5327 | Yes | ||
| 169 | MORC2 | 2370750 | 17149 | -0.263 | -0.5231 | Yes | ||
| 170 | RNF11 | 3990068 5080458 | 17385 | -0.327 | -0.5228 | Yes | ||
| 171 | TOB1 | 4150138 | 17435 | -0.340 | -0.5118 | Yes | ||
| 172 | CDC42 | 1240168 3440278 4480519 5290162 | 17630 | -0.401 | -0.5063 | Yes | ||
| 173 | PDCD10 | 4760500 | 17634 | -0.404 | -0.4903 | Yes | ||
| 174 | POLS | 5890438 | 17665 | -0.415 | -0.4753 | Yes | ||
| 175 | DDX3X | 2190020 | 17682 | -0.421 | -0.4593 | Yes | ||
| 176 | EPC2 | 2470095 | 17729 | -0.437 | -0.4443 | Yes | ||
| 177 | DPH2 | 2100402 3060039 | 17736 | -0.439 | -0.4270 | Yes | ||
| 178 | AHCYL1 | 540609 | 17737 | -0.440 | -0.4094 | Yes | ||
| 179 | SGPP1 | 3930707 | 17740 | -0.441 | -0.3919 | Yes | ||
| 180 | PABPC1 | 2650180 2690253 6020632 1990270 | 17866 | -0.488 | -0.3791 | Yes | ||
| 181 | MBNL2 | 3450707 | 17905 | -0.509 | -0.3608 | Yes | ||
| 182 | BCL11B | 2680673 | 17919 | -0.514 | -0.3409 | Yes | ||
| 183 | ZHX1 | 2100110 2120286 | 18084 | -0.606 | -0.3256 | Yes | ||
| 184 | MTPN | 3940167 | 18102 | -0.614 | -0.3019 | Yes | ||
| 185 | RIOK3 | 1980487 4480619 7040026 | 18103 | -0.614 | -0.2773 | Yes | ||
| 186 | CLK4 | 3440484 3610156 | 18137 | -0.636 | -0.2536 | Yes | ||
| 187 | APRIN | 2810050 | 18163 | -0.652 | -0.2289 | Yes | ||
| 188 | EED | 2320373 | 18198 | -0.671 | -0.2038 | Yes | ||
| 189 | NASP | 2260139 2940369 5130707 | 18205 | -0.676 | -0.1771 | Yes | ||
| 190 | SUMO1 | 1990364 2760338 6380195 | 18227 | -0.696 | -0.1504 | Yes | ||
| 191 | SRPK2 | 6380341 | 18312 | -0.780 | -0.1237 | Yes | ||
| 192 | PARG | 50139 | 18379 | -0.861 | -0.0928 | Yes | ||
| 193 | MBNL1 | 2640762 7100048 | 18381 | -0.865 | -0.0582 | Yes | ||
| 194 | EIF2S2 | 770095 2810487 | 18392 | -0.876 | -0.0237 | Yes | ||
| 195 | APLP2 | 3610037 | 18405 | -0.895 | 0.0115 | Yes |

