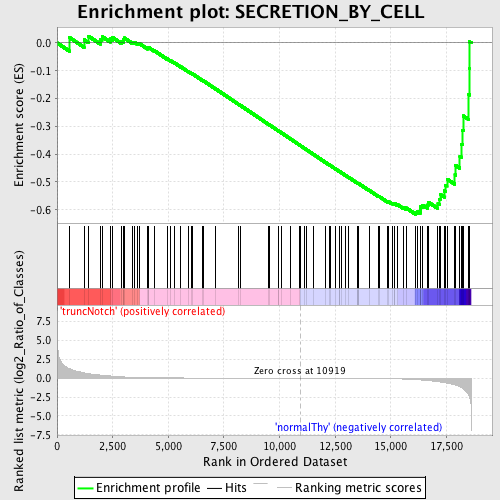
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | SECRETION_BY_CELL |
| Enrichment Score (ES) | -0.6168234 |
| Normalized Enrichment Score (NES) | -1.4954687 |
| Nominal p-value | 0.005221932 |
| FDR q-value | 0.5825909 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CCL8 | 3870010 | 551 | 1.265 | 0.0195 | No | ||
| 2 | APOA1 | 110152 | 1214 | 0.706 | 0.0113 | No | ||
| 3 | RAB3A | 3940288 | 1406 | 0.602 | 0.0244 | No | ||
| 4 | NAPA | 1780537 5720102 | 1940 | 0.418 | 0.0120 | No | ||
| 5 | ARL4D | 1570154 | 2018 | 0.390 | 0.0230 | No | ||
| 6 | CADM1 | 6620128 | 2389 | 0.294 | 0.0145 | No | ||
| 7 | ERGIC3 | 1400075 4780047 | 2499 | 0.265 | 0.0189 | No | ||
| 8 | SNAP23 | 2480722 5390315 | 2887 | 0.183 | 0.0052 | No | ||
| 9 | SCRN1 | 6040025 6580019 | 2972 | 0.169 | 0.0072 | No | ||
| 10 | CCL3 | 2810092 | 2985 | 0.168 | 0.0131 | No | ||
| 11 | COPZ1 | 6940100 | 3014 | 0.164 | 0.0180 | No | ||
| 12 | SEPT5 | 460725 | 3380 | 0.119 | 0.0029 | No | ||
| 13 | COPE | 2340088 2640450 6620279 | 3483 | 0.110 | 0.0017 | No | ||
| 14 | SCG2 | 5670438 130671 | 3599 | 0.100 | -0.0006 | No | ||
| 15 | VTI1B | 3140364 6510497 | 3710 | 0.092 | -0.0030 | No | ||
| 16 | NMUR2 | 1580717 | 4066 | 0.073 | -0.0193 | No | ||
| 17 | ARFGEF1 | 6760494 | 4109 | 0.071 | -0.0188 | No | ||
| 18 | AKAP3 | 5890471 | 4115 | 0.071 | -0.0163 | No | ||
| 19 | KIF1C | 2480484 | 4379 | 0.059 | -0.0282 | No | ||
| 20 | BACE2 | 4060170 5690100 6220133 | 4945 | 0.042 | -0.0570 | No | ||
| 21 | APOA2 | 6510364 6860411 | 5114 | 0.039 | -0.0646 | No | ||
| 22 | OPTN | 3940041 | 5269 | 0.036 | -0.0715 | No | ||
| 23 | CANX | 4760402 | 5561 | 0.030 | -0.0860 | No | ||
| 24 | RIMS1 | 5670022 | 5901 | 0.025 | -0.1034 | No | ||
| 25 | AP3B2 | 4670605 | 6031 | 0.023 | -0.1094 | No | ||
| 26 | CIDEA | 4560020 | 6072 | 0.023 | -0.1107 | No | ||
| 27 | GRM4 | 6960092 | 6531 | 0.018 | -0.1347 | No | ||
| 28 | VAMP3 | 7100050 | 6559 | 0.018 | -0.1355 | No | ||
| 29 | NLRC4 | 5570332 | 7097 | 0.014 | -0.1639 | No | ||
| 30 | LIN7A | 4540707 5270369 | 8147 | 0.009 | -0.2202 | No | ||
| 31 | PPY | 2340373 | 8234 | 0.008 | -0.2245 | No | ||
| 32 | NLRP2 | 580750 | 9523 | 0.004 | -0.2939 | No | ||
| 33 | CADPS | 2320181 | 9555 | 0.004 | -0.2954 | No | ||
| 34 | NLRP3 | 1980064 4010180 | 9931 | 0.003 | -0.3155 | No | ||
| 35 | BET1 | 4010725 | 9934 | 0.003 | -0.3155 | No | ||
| 36 | ABCA1 | 6290156 | 10074 | 0.002 | -0.3229 | No | ||
| 37 | COPB1 | 5390037 | 10505 | 0.001 | -0.3461 | No | ||
| 38 | EXOC5 | 7000711 | 10889 | 0.000 | -0.3667 | No | ||
| 39 | SEC22A | 3800162 3840168 5220164 5290632 | 10932 | -0.000 | -0.3690 | No | ||
| 40 | DNAJC1 | 6650066 | 11120 | -0.001 | -0.3791 | No | ||
| 41 | GLMN | 5290563 5890239 | 11202 | -0.001 | -0.3834 | No | ||
| 42 | PKDREJ | 4200088 | 11517 | -0.002 | -0.4003 | No | ||
| 43 | STX16 | 70315 | 12083 | -0.004 | -0.4307 | No | ||
| 44 | SYN3 | 3290427 | 12247 | -0.004 | -0.4393 | No | ||
| 45 | NLGN1 | 5670278 | 12288 | -0.005 | -0.4413 | No | ||
| 46 | TXNDC4 | 450537 | 12515 | -0.006 | -0.4532 | No | ||
| 47 | CPLX1 | 2510735 | 12680 | -0.006 | -0.4618 | No | ||
| 48 | SYT1 | 840364 | 12700 | -0.006 | -0.4626 | No | ||
| 49 | CPLX2 | 6980538 | 12786 | -0.007 | -0.4669 | No | ||
| 50 | ACHE | 5290750 | 12979 | -0.008 | -0.4770 | No | ||
| 51 | NMUR1 | 1500403 | 13099 | -0.009 | -0.4831 | No | ||
| 52 | CARTPT | 6660687 | 13495 | -0.012 | -0.5039 | No | ||
| 53 | LTBP2 | 4850039 | 13501 | -0.012 | -0.5037 | No | ||
| 54 | SNCAIP | 4010551 | 13539 | -0.013 | -0.5052 | No | ||
| 55 | COG2 | 5900129 | 13554 | -0.013 | -0.5055 | No | ||
| 56 | SEC22B | 3190278 | 14030 | -0.019 | -0.5304 | No | ||
| 57 | CRTAM | 4480110 | 14432 | -0.029 | -0.5509 | No | ||
| 58 | RPH3AL | 70110 | 14481 | -0.031 | -0.5523 | No | ||
| 59 | NOD2 | 2510050 | 14872 | -0.050 | -0.5714 | No | ||
| 60 | SYTL4 | 3060066 | 14894 | -0.051 | -0.5705 | No | ||
| 61 | HRH3 | 6180040 | 15059 | -0.061 | -0.5769 | No | ||
| 62 | SEC22C | 1050600 6760050 | 15155 | -0.068 | -0.5794 | No | ||
| 63 | LMAN1 | 3420068 | 15178 | -0.070 | -0.5779 | No | ||
| 64 | RAB26 | 5910025 | 15291 | -0.079 | -0.5809 | No | ||
| 65 | GOSR2 | 1850059 1980687 4590184 | 15574 | -0.109 | -0.5918 | No | ||
| 66 | COG7 | 360133 | 15689 | -0.121 | -0.5933 | No | ||
| 67 | ARFGEF2 | 5550500 | 16126 | -0.181 | -0.6098 | Yes | ||
| 68 | STX18 | 4760706 | 16181 | -0.193 | -0.6052 | Yes | ||
| 69 | FOXP3 | 940707 1850692 | 16316 | -0.219 | -0.6039 | Yes | ||
| 70 | SCAMP3 | 2360707 | 16337 | -0.223 | -0.5963 | Yes | ||
| 71 | TXNDC1 | 520398 | 16345 | -0.225 | -0.5879 | Yes | ||
| 72 | ARFGAP3 | 3390717 | 16429 | -0.244 | -0.5828 | Yes | ||
| 73 | COPG2 | 2370152 | 16647 | -0.306 | -0.5826 | Yes | ||
| 74 | SCAMP2 | 430114 | 16677 | -0.314 | -0.5720 | Yes | ||
| 75 | KCNMB4 | 3290347 | 17118 | -0.459 | -0.5778 | Yes | ||
| 76 | PYCARD | 2100750 | 17196 | -0.490 | -0.5629 | Yes | ||
| 77 | ARFIP1 | 3130601 | 17218 | -0.499 | -0.5446 | Yes | ||
| 78 | GOSR1 | 730279 | 17401 | -0.579 | -0.5319 | Yes | ||
| 79 | NRBP1 | 1740750 | 17465 | -0.617 | -0.5112 | Yes | ||
| 80 | COG3 | 3840746 | 17526 | -0.644 | -0.4894 | Yes | ||
| 81 | COPB2 | 610154 | 17883 | -0.876 | -0.4745 | Yes | ||
| 82 | SNAP29 | 6380239 | 17910 | -0.907 | -0.4406 | Yes | ||
| 83 | RAB2A | 2450358 | 18105 | -1.119 | -0.4074 | Yes | ||
| 84 | RAB14 | 6860139 | 18184 | -1.233 | -0.3636 | Yes | ||
| 85 | SCIN | 5420180 | 18226 | -1.318 | -0.3145 | Yes | ||
| 86 | STX7 | 5910484 | 18259 | -1.384 | -0.2623 | Yes | ||
| 87 | ZW10 | 2900735 3520687 | 18502 | -2.294 | -0.1860 | Yes | ||
| 88 | LMAN2L | 50079 6590100 6650338 | 18532 | -2.438 | -0.0926 | Yes | ||
| 89 | CCL5 | 3710397 | 18540 | -2.493 | 0.0041 | Yes |

