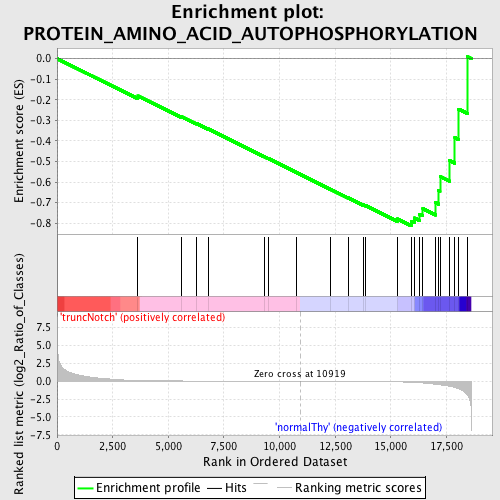
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | PROTEIN_AMINO_ACID_AUTOPHOSPHORYLATION |
| Enrichment Score (ES) | -0.813261 |
| Normalized Enrichment Score (NES) | -1.5495471 |
| Nominal p-value | 0.002352941 |
| FDR q-value | 0.56780183 |
| FWER p-Value | 0.963 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MAP3K10 | 3360168 4850180 | 3634 | 0.098 | -0.1815 | No | ||
| 2 | TTN | 2320161 4670056 6550026 | 5592 | 0.030 | -0.2825 | No | ||
| 3 | MAP3K9 | 3290707 | 6252 | 0.021 | -0.3150 | No | ||
| 4 | TNK1 | 4730279 | 6791 | 0.016 | -0.3416 | No | ||
| 5 | AKT1 | 5290746 | 9329 | 0.005 | -0.4774 | No | ||
| 6 | MAP3K13 | 3190017 | 9487 | 0.004 | -0.4852 | No | ||
| 7 | MYO3A | 5290494 | 10754 | 0.000 | -0.5533 | No | ||
| 8 | MAP3K12 | 2470373 6420162 | 12294 | -0.005 | -0.6354 | No | ||
| 9 | CDKL5 | 2450070 | 13081 | -0.009 | -0.6764 | No | ||
| 10 | INSR | 1190504 | 13754 | -0.015 | -0.7104 | No | ||
| 11 | PAK1 | 4540315 | 13867 | -0.017 | -0.7141 | No | ||
| 12 | UHMK1 | 3850670 6450064 | 15285 | -0.079 | -0.7791 | No | ||
| 13 | LMTK2 | 6650692 | 15922 | -0.152 | -0.7917 | Yes | ||
| 14 | IGF1R | 3360494 | 16054 | -0.171 | -0.7744 | Yes | ||
| 15 | CAMKK2 | 5270047 | 16291 | -0.214 | -0.7566 | Yes | ||
| 16 | CRKRS | 510537 2510551 | 16441 | -0.247 | -0.7293 | Yes | ||
| 17 | MAP3K11 | 7000039 | 17016 | -0.419 | -0.7005 | Yes | ||
| 18 | EIF2AK1 | 2470301 | 17148 | -0.472 | -0.6403 | Yes | ||
| 19 | PAK2 | 360438 7050068 | 17211 | -0.496 | -0.5729 | Yes | ||
| 20 | EIF2AK3 | 4280161 | 17636 | -0.700 | -0.4959 | Yes | ||
| 21 | MAP3K3 | 610685 | 17879 | -0.873 | -0.3844 | Yes | ||
| 22 | DYRK1A | 3190181 | 18053 | -1.036 | -0.2461 | Yes | ||
| 23 | STK4 | 2640152 | 18455 | -1.939 | 0.0087 | Yes |

