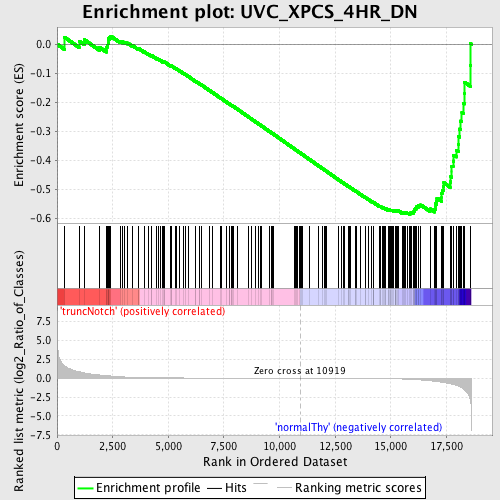
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | UVC_XPCS_4HR_DN |
| Enrichment Score (ES) | -0.5859355 |
| Normalized Enrichment Score (NES) | -1.5454402 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.28492197 |
| FWER p-Value | 0.99 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATXN2 | 3940017 4810242 | 322 | 1.643 | 0.0246 | No | ||
| 2 | MTF2 | 50333 | 990 | 0.856 | 0.0103 | No | ||
| 3 | RCOR1 | 5360717 | 1229 | 0.699 | 0.0153 | No | ||
| 4 | PRR3 | 3440112 | 1903 | 0.427 | -0.0103 | No | ||
| 5 | PTK2 | 1780148 | 2213 | 0.339 | -0.0184 | No | ||
| 6 | MYST3 | 5270500 | 2215 | 0.339 | -0.0098 | No | ||
| 7 | EIF2C2 | 770451 | 2248 | 0.331 | -0.0030 | No | ||
| 8 | MTAP | 2120136 | 2287 | 0.321 | 0.0031 | No | ||
| 9 | HIRA | 4480609 5270167 | 2299 | 0.318 | 0.0107 | No | ||
| 10 | BHLHB2 | 7040603 | 2302 | 0.317 | 0.0187 | No | ||
| 11 | LHFPL2 | 450020 | 2361 | 0.301 | 0.0232 | No | ||
| 12 | ZNF148 | 6420403 | 2414 | 0.289 | 0.0278 | No | ||
| 13 | MICAL3 | 6040333 | 2835 | 0.190 | 0.0099 | No | ||
| 14 | RBM16 | 6290286 6400181 | 2946 | 0.174 | 0.0084 | No | ||
| 15 | PBX3 | 1300424 3710577 6180575 | 3041 | 0.161 | 0.0074 | No | ||
| 16 | AKAP9 | 510184 2060195 2470136 4280048 | 3168 | 0.143 | 0.0042 | No | ||
| 17 | EXT1 | 4540603 | 3377 | 0.119 | -0.0040 | No | ||
| 18 | AKAP10 | 2630504 5910577 | 3649 | 0.097 | -0.0162 | No | ||
| 19 | ANKS1A | 50148 | 3660 | 0.096 | -0.0143 | No | ||
| 20 | SHB | 4920494 | 3909 | 0.080 | -0.0257 | No | ||
| 21 | ARFGEF1 | 6760494 | 4109 | 0.071 | -0.0347 | No | ||
| 22 | DLC1 | 1090632 6450594 | 4227 | 0.066 | -0.0394 | No | ||
| 23 | SOCS5 | 3830398 7100093 | 4232 | 0.065 | -0.0379 | No | ||
| 24 | HOXB2 | 6450592 | 4477 | 0.056 | -0.0497 | No | ||
| 25 | SWAP70 | 2810577 2850739 5130594 | 4547 | 0.054 | -0.0521 | No | ||
| 26 | CDK8 | 1050113 1190601 1780358 6110102 | 4646 | 0.051 | -0.0561 | No | ||
| 27 | DOCK4 | 5910102 | 4757 | 0.048 | -0.0608 | No | ||
| 28 | CDYL | 4200142 4730397 6060400 | 4786 | 0.047 | -0.0611 | No | ||
| 29 | TUBGCP3 | 4920563 | 4813 | 0.046 | -0.0614 | No | ||
| 30 | NFIB | 460450 | 4827 | 0.045 | -0.0609 | No | ||
| 31 | TLE1 | 540390 1580309 3710019 | 5076 | 0.040 | -0.0734 | No | ||
| 32 | ACVR1 | 6840671 | 5119 | 0.039 | -0.0746 | No | ||
| 33 | FGF2 | 3450433 | 5133 | 0.038 | -0.0744 | No | ||
| 34 | KIF2A | 3990286 6130575 | 5329 | 0.035 | -0.0840 | No | ||
| 35 | CTGF | 4540577 | 5358 | 0.034 | -0.0847 | No | ||
| 36 | STK3 | 7100427 | 5484 | 0.032 | -0.0907 | No | ||
| 37 | EGFR | 4920138 6480521 | 5695 | 0.028 | -0.1013 | No | ||
| 38 | BMPR1A | 2190193 4060603 | 5766 | 0.027 | -0.1044 | No | ||
| 39 | WNT5A | 840685 3120152 | 5922 | 0.025 | -0.1122 | No | ||
| 40 | TSPAN5 | 6770019 | 6207 | 0.021 | -0.1270 | No | ||
| 41 | BICD1 | 110128 2940324 | 6229 | 0.021 | -0.1276 | No | ||
| 42 | BCAR3 | 1660717 | 6394 | 0.019 | -0.1360 | No | ||
| 43 | CENPE | 2850022 | 6417 | 0.019 | -0.1367 | No | ||
| 44 | CD2AP | 1940369 | 6506 | 0.019 | -0.1410 | No | ||
| 45 | MID1 | 4070114 4760458 | 6849 | 0.016 | -0.1592 | No | ||
| 46 | CENTG2 | 4210053 | 6979 | 0.015 | -0.1658 | No | ||
| 47 | DDX10 | 520746 | 7330 | 0.013 | -0.1844 | No | ||
| 48 | PLEKHC1 | 2650066 | 7384 | 0.012 | -0.1870 | No | ||
| 49 | SRGAP2 | 1050092 1230403 3830066 6130215 | 7597 | 0.011 | -0.1982 | No | ||
| 50 | SOS1 | 7050338 | 7726 | 0.010 | -0.2049 | No | ||
| 51 | ARHGAP12 | 1050181 | 7751 | 0.010 | -0.2059 | No | ||
| 52 | RNF13 | 2370021 | 7843 | 0.010 | -0.2106 | No | ||
| 53 | CBLB | 3520465 4670348 | 7854 | 0.010 | -0.2109 | No | ||
| 54 | FBXL7 | 6620711 | 7883 | 0.010 | -0.2121 | No | ||
| 55 | CTBP2 | 430309 3710079 | 7928 | 0.010 | -0.2143 | No | ||
| 56 | PAFAH1B1 | 4230333 6420121 6450066 | 8128 | 0.009 | -0.2248 | No | ||
| 57 | ACSL3 | 3140195 | 8607 | 0.007 | -0.2506 | No | ||
| 58 | SCHIP1 | 4560152 | 8735 | 0.006 | -0.2573 | No | ||
| 59 | NCK1 | 6200575 6510050 | 8930 | 0.006 | -0.2677 | No | ||
| 60 | TIPARP | 50397 | 9033 | 0.005 | -0.2731 | No | ||
| 61 | SAMD4A | 1170717 | 9135 | 0.005 | -0.2784 | No | ||
| 62 | STRN3 | 1450093 6200685 | 9199 | 0.005 | -0.2817 | No | ||
| 63 | ZCCHC11 | 3390731 5360039 | 9552 | 0.004 | -0.3007 | No | ||
| 64 | EHBP1 | 430348 | 9617 | 0.004 | -0.3040 | No | ||
| 65 | RGS7 | 3390044 | 9640 | 0.004 | -0.3051 | No | ||
| 66 | TRIM2 | 430546 1170286 | 9661 | 0.004 | -0.3061 | No | ||
| 67 | KLF10 | 4850056 | 9725 | 0.003 | -0.3095 | No | ||
| 68 | RNGTT | 780152 780746 1090471 | 10669 | 0.001 | -0.3606 | No | ||
| 69 | FGF5 | 5270204 | 10715 | 0.001 | -0.3630 | No | ||
| 70 | MYC | 380541 4670170 | 10764 | 0.000 | -0.3656 | No | ||
| 71 | BLM | 520619 5570170 | 10797 | 0.000 | -0.3673 | No | ||
| 72 | TAF4 | 1450170 | 10913 | 0.000 | -0.3735 | No | ||
| 73 | IL1RAP | 50546 450711 1580397 3940301 5890181 6370373 | 10939 | -0.000 | -0.3749 | No | ||
| 74 | SATB2 | 2470181 | 10987 | -0.000 | -0.3774 | No | ||
| 75 | CENPC1 | 610273 | 11038 | -0.000 | -0.3801 | No | ||
| 76 | DAPK1 | 5570010 | 11348 | -0.001 | -0.3969 | No | ||
| 77 | IRAK1BP1 | 610037 | 11746 | -0.003 | -0.4183 | No | ||
| 78 | STARD13 | 540722 | 11912 | -0.003 | -0.4272 | No | ||
| 79 | GNAI1 | 4560390 | 12039 | -0.004 | -0.4339 | No | ||
| 80 | PVRL3 | 1400324 4200593 6650128 | 12081 | -0.004 | -0.4360 | No | ||
| 81 | NUP153 | 7000452 | 12121 | -0.004 | -0.4380 | No | ||
| 82 | F3 | 2940180 | 12655 | -0.006 | -0.4668 | No | ||
| 83 | TERF1 | 2340113 | 12802 | -0.007 | -0.4745 | No | ||
| 84 | VEGFC | 5910494 | 12885 | -0.007 | -0.4788 | No | ||
| 85 | ZFYVE9 | 6980288 7100017 | 12909 | -0.008 | -0.4798 | No | ||
| 86 | GNAQ | 430670 4210131 5900736 | 13080 | -0.009 | -0.4888 | No | ||
| 87 | FBXW11 | 6450632 | 13100 | -0.009 | -0.4896 | No | ||
| 88 | ARHGEF7 | 1690441 | 13144 | -0.009 | -0.4917 | No | ||
| 89 | USP15 | 610592 3520504 | 13188 | -0.009 | -0.4938 | No | ||
| 90 | TGFB2 | 4920292 | 13398 | -0.011 | -0.5049 | No | ||
| 91 | E2F5 | 5860575 | 13405 | -0.011 | -0.5049 | No | ||
| 92 | IL6 | 380133 | 13456 | -0.012 | -0.5073 | No | ||
| 93 | NRIP1 | 3190039 | 13626 | -0.014 | -0.5161 | No | ||
| 94 | TOPBP1 | 6020333 | 13849 | -0.016 | -0.5277 | No | ||
| 95 | PUM2 | 4200441 5910446 | 13875 | -0.017 | -0.5287 | No | ||
| 96 | GLRB | 2510433 4280736 | 14000 | -0.019 | -0.5349 | No | ||
| 97 | CLASP2 | 2510139 | 14152 | -0.021 | -0.5425 | No | ||
| 98 | ZHX3 | 460615 | 14207 | -0.023 | -0.5449 | No | ||
| 99 | FNDC3A | 2690097 3290044 | 14220 | -0.023 | -0.5449 | No | ||
| 100 | PIP5K3 | 5360112 | 14234 | -0.023 | -0.5451 | No | ||
| 101 | EPHA4 | 460750 | 14509 | -0.032 | -0.5591 | No | ||
| 102 | IFRD1 | 4590215 | 14518 | -0.032 | -0.5587 | No | ||
| 103 | CYR61 | 1240408 5290026 4120452 6550008 | 14530 | -0.033 | -0.5585 | No | ||
| 104 | PTPRK | 1450433 2640142 | 14612 | -0.036 | -0.5619 | No | ||
| 105 | PARG | 50139 | 14648 | -0.038 | -0.5628 | No | ||
| 106 | SEC24B | 460717 3060040 | 14706 | -0.041 | -0.5649 | No | ||
| 107 | ANKRD17 | 2120402 3850035 6400044 | 14765 | -0.044 | -0.5669 | No | ||
| 108 | CREB1 | 1500717 2230358 3610600 6550601 | 14774 | -0.045 | -0.5662 | No | ||
| 109 | PTPN12 | 2030309 6020725 6130746 6290609 | 14883 | -0.051 | -0.5707 | No | ||
| 110 | STAM | 3850008 4760441 | 14899 | -0.052 | -0.5702 | No | ||
| 111 | MALT1 | 4670292 | 14919 | -0.053 | -0.5699 | No | ||
| 112 | ZZZ3 | 780133 2320239 2640452 5080706 6550341 | 14970 | -0.055 | -0.5712 | No | ||
| 113 | APRIN | 2810050 | 14986 | -0.056 | -0.5706 | No | ||
| 114 | THRAP1 | 2470121 | 15024 | -0.059 | -0.5711 | No | ||
| 115 | TRIM33 | 580619 2230280 3990433 6200747 | 15075 | -0.063 | -0.5722 | No | ||
| 116 | PHLPP | 3440563 | 15138 | -0.067 | -0.5738 | No | ||
| 117 | CNOT2 | 520546 2060397 4610543 5550113 | 15191 | -0.071 | -0.5748 | No | ||
| 118 | MARCH7 | 3800020 6620168 | 15198 | -0.071 | -0.5733 | No | ||
| 119 | PCTK2 | 5700673 | 15231 | -0.073 | -0.5732 | No | ||
| 120 | MTMR3 | 1050014 4920180 | 15258 | -0.076 | -0.5727 | No | ||
| 121 | DNMBP | 2810368 6900403 | 15286 | -0.079 | -0.5721 | No | ||
| 122 | MYST4 | 1400563 2570687 3360458 6840402 | 15322 | -0.082 | -0.5719 | No | ||
| 123 | TGFBR3 | 5290577 | 15505 | -0.101 | -0.5792 | No | ||
| 124 | MYO10 | 3830576 5570092 | 15591 | -0.111 | -0.5810 | No | ||
| 125 | REV3L | 1090717 | 15593 | -0.111 | -0.5782 | No | ||
| 126 | SLC25A12 | 7400731 | 15655 | -0.117 | -0.5785 | No | ||
| 127 | FYN | 2100468 4760520 4850687 | 15761 | -0.130 | -0.5809 | No | ||
| 128 | TCF7L2 | 2190348 5220113 | 15855 | -0.142 | -0.5823 | Yes | ||
| 129 | CNOT4 | 2320242 6760706 | 15894 | -0.148 | -0.5806 | Yes | ||
| 130 | SOS2 | 2060722 | 15919 | -0.151 | -0.5780 | Yes | ||
| 131 | PPP1R12A | 4070066 4590162 | 16004 | -0.164 | -0.5784 | Yes | ||
| 132 | BTRC | 3170131 | 16035 | -0.169 | -0.5757 | Yes | ||
| 133 | IGF1R | 3360494 | 16054 | -0.171 | -0.5723 | Yes | ||
| 134 | ORC5L | 1940133 1940711 | 16076 | -0.174 | -0.5690 | Yes | ||
| 135 | KLF6 | 110541 5360026 | 16103 | -0.178 | -0.5659 | Yes | ||
| 136 | ATF2 | 2260348 2480441 | 16133 | -0.183 | -0.5627 | Yes | ||
| 137 | ZNF292 | 840736 1500390 | 16169 | -0.191 | -0.5598 | Yes | ||
| 138 | UPF2 | 4810471 | 16228 | -0.201 | -0.5578 | Yes | ||
| 139 | ZHX2 | 2900452 | 16244 | -0.206 | -0.5533 | Yes | ||
| 140 | SMURF2 | 1940161 | 16348 | -0.226 | -0.5531 | Yes | ||
| 141 | BARD1 | 3170348 | 16760 | -0.336 | -0.5668 | Yes | ||
| 142 | CENPA | 5080154 | 16958 | -0.400 | -0.5673 | Yes | ||
| 143 | CHD1 | 4590048 | 17013 | -0.419 | -0.5595 | Yes | ||
| 144 | CREBBP | 5690035 7040050 | 17020 | -0.421 | -0.5491 | Yes | ||
| 145 | WDR37 | 2630519 6840347 | 17051 | -0.431 | -0.5397 | Yes | ||
| 146 | CENTD1 | 2570592 | 17071 | -0.437 | -0.5295 | Yes | ||
| 147 | MECP2 | 1940450 | 17281 | -0.523 | -0.5275 | Yes | ||
| 148 | ITSN2 | 780484 | 17287 | -0.525 | -0.5143 | Yes | ||
| 149 | THRAP2 | 4920600 | 17307 | -0.533 | -0.5018 | Yes | ||
| 150 | NCOA3 | 4540195 | 17359 | -0.562 | -0.4902 | Yes | ||
| 151 | RBM39 | 4230458 6450280 | 17384 | -0.573 | -0.4768 | Yes | ||
| 152 | RRAS2 | 6290170 | 17666 | -0.719 | -0.4737 | Yes | ||
| 153 | RASA1 | 1240315 | 17696 | -0.741 | -0.4563 | Yes | ||
| 154 | TIAM1 | 5420288 | 17739 | -0.768 | -0.4390 | Yes | ||
| 155 | TLK2 | 1170161 | 17740 | -0.768 | -0.4193 | Yes | ||
| 156 | SRPK2 | 6380341 | 17799 | -0.805 | -0.4019 | Yes | ||
| 157 | TLK1 | 7100605 | 17823 | -0.828 | -0.3820 | Yes | ||
| 158 | SPEN | 2060041 | 17938 | -0.930 | -0.3644 | Yes | ||
| 159 | APPBP2 | 5130215 | 18043 | -1.031 | -0.3437 | Yes | ||
| 160 | DYRK1A | 3190181 | 18053 | -1.036 | -0.3177 | Yes | ||
| 161 | NCOA1 | 3610438 | 18091 | -1.092 | -0.2918 | Yes | ||
| 162 | FCHSD2 | 5720092 | 18137 | -1.173 | -0.2642 | Yes | ||
| 163 | MARK3 | 2370450 | 18196 | -1.253 | -0.2354 | Yes | ||
| 164 | RUNX1 | 3840711 | 18253 | -1.370 | -0.2034 | Yes | ||
| 165 | KLF7 | 50390 | 18299 | -1.477 | -0.1681 | Yes | ||
| 166 | UGCG | 3420053 | 18307 | -1.492 | -0.1303 | Yes | ||
| 167 | TLE4 | 1450064 1500519 | 18569 | -2.751 | -0.0742 | Yes | ||
| 168 | CUTL1 | 1570113 1770091 1850273 2100286 5340605 | 18583 | -2.998 | 0.0018 | Yes |

