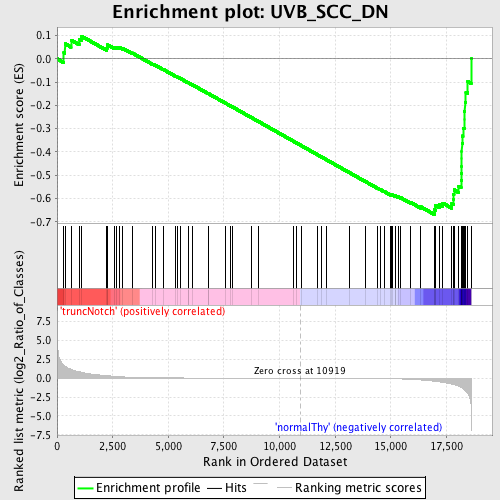
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | UVB_SCC_DN |
| Enrichment Score (ES) | -0.6687097 |
| Normalized Enrichment Score (NES) | -1.5618856 |
| Nominal p-value | 0.0025316456 |
| FDR q-value | 0.27409276 |
| FWER p-Value | 0.981 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DDIT4 | 2570408 | 305 | 1.678 | 0.0268 | No | ||
| 2 | SEPT8 | 4610463 | 354 | 1.587 | 0.0650 | No | ||
| 3 | DUSP4 | 2690044 2850593 | 625 | 1.163 | 0.0804 | No | ||
| 4 | MTF2 | 50333 | 990 | 0.856 | 0.0829 | No | ||
| 5 | KIFC1 | 2570332 2480725 | 1092 | 0.785 | 0.0976 | No | ||
| 6 | MYST3 | 5270500 | 2215 | 0.339 | 0.0459 | No | ||
| 7 | EIF2C2 | 770451 | 2248 | 0.331 | 0.0527 | No | ||
| 8 | RPP38 | 2450164 | 2270 | 0.326 | 0.0599 | No | ||
| 9 | TRAF4 | 3060041 4920528 6980286 | 2583 | 0.244 | 0.0494 | No | ||
| 10 | AURKA | 780537 | 2673 | 0.224 | 0.0504 | No | ||
| 11 | FZD2 | 6480154 | 2791 | 0.197 | 0.0491 | No | ||
| 12 | PHC2 | 1430433 | 2938 | 0.174 | 0.0458 | No | ||
| 13 | EXT1 | 4540603 | 3377 | 0.119 | 0.0252 | No | ||
| 14 | CSTF1 | 4610129 | 4303 | 0.062 | -0.0231 | No | ||
| 15 | YTHDF3 | 4540632 4670717 7040609 | 4429 | 0.057 | -0.0283 | No | ||
| 16 | CDYL | 4200142 4730397 6060400 | 4786 | 0.047 | -0.0463 | No | ||
| 17 | KIF2A | 3990286 6130575 | 5329 | 0.035 | -0.0747 | No | ||
| 18 | ZC3H8 | 3360524 | 5414 | 0.033 | -0.0784 | No | ||
| 19 | TRAM2 | 3940112 | 5565 | 0.030 | -0.0857 | No | ||
| 20 | ERF | 110551 1770278 | 5889 | 0.025 | -0.1024 | No | ||
| 21 | ADORA2B | 6860253 | 5894 | 0.025 | -0.1020 | No | ||
| 22 | TUSC2 | 1580253 | 6091 | 0.022 | -0.1120 | No | ||
| 23 | POGZ | 870348 7040286 | 6788 | 0.016 | -0.1491 | No | ||
| 24 | RRS1 | 4560551 | 6818 | 0.016 | -0.1503 | No | ||
| 25 | E2F3 | 50162 460180 | 7560 | 0.011 | -0.1899 | No | ||
| 26 | AMD1 | 6290128 | 7794 | 0.010 | -0.2023 | No | ||
| 27 | PMM2 | 670673 2570044 2690093 4540068 6450056 | 7895 | 0.010 | -0.2074 | No | ||
| 28 | SCHIP1 | 4560152 | 8735 | 0.006 | -0.2525 | No | ||
| 29 | TIPARP | 50397 | 9033 | 0.005 | -0.2684 | No | ||
| 30 | HMGA2 | 2940121 3390647 5130279 6400136 | 10643 | 0.001 | -0.3551 | No | ||
| 31 | MYC | 380541 4670170 | 10764 | 0.000 | -0.3616 | No | ||
| 32 | MFAP1 | 6660092 | 10988 | -0.000 | -0.3736 | No | ||
| 33 | SHOC2 | 1340064 1570546 | 11720 | -0.003 | -0.4129 | No | ||
| 34 | FOXF2 | 1240091 | 11898 | -0.003 | -0.4224 | No | ||
| 35 | NUP153 | 7000452 | 12121 | -0.004 | -0.4343 | No | ||
| 36 | ARHGEF7 | 1690441 | 13144 | -0.009 | -0.4892 | No | ||
| 37 | DDX3Y | 1580278 4200519 | 13858 | -0.016 | -0.5272 | No | ||
| 38 | BAG5 | 2230368 6420537 | 14390 | -0.028 | -0.5551 | No | ||
| 39 | CYR61 | 1240408 5290026 4120452 6550008 | 14530 | -0.033 | -0.5618 | No | ||
| 40 | WEE1 | 3390070 | 14710 | -0.041 | -0.5703 | No | ||
| 41 | BLCAP | 3990253 | 14992 | -0.057 | -0.5840 | No | ||
| 42 | BRD8 | 1170463 2900075 4050397 6590332 | 15016 | -0.058 | -0.5838 | No | ||
| 43 | COIL | 4480408 | 15062 | -0.062 | -0.5846 | No | ||
| 44 | FADD | 2360593 4210692 6520019 | 15189 | -0.071 | -0.5896 | No | ||
| 45 | CNOT2 | 520546 2060397 4610543 5550113 | 15191 | -0.071 | -0.5878 | No | ||
| 46 | SIAH2 | 7210672 | 15325 | -0.083 | -0.5929 | No | ||
| 47 | BAIAP2 | 460347 4730600 | 15441 | -0.094 | -0.5966 | No | ||
| 48 | CNOT4 | 2320242 6760706 | 15894 | -0.148 | -0.6172 | No | ||
| 49 | CASP8 | 4850154 | 16340 | -0.224 | -0.6354 | No | ||
| 50 | CENPA | 5080154 | 16958 | -0.400 | -0.6584 | Yes | ||
| 51 | TNFAIP8 | 2760551 | 16962 | -0.402 | -0.6482 | Yes | ||
| 52 | CHD1 | 4590048 | 17013 | -0.419 | -0.6401 | Yes | ||
| 53 | HMGCR | 2630242 | 17015 | -0.419 | -0.6294 | Yes | ||
| 54 | NAGK | 730041 3170086 5890053 | 17193 | -0.489 | -0.6263 | Yes | ||
| 55 | RYBP | 60746 | 17335 | -0.550 | -0.6198 | Yes | ||
| 56 | TLK2 | 1170161 | 17740 | -0.768 | -0.6218 | Yes | ||
| 57 | GATA3 | 6130068 | 17815 | -0.824 | -0.6045 | Yes | ||
| 58 | TLK1 | 7100605 | 17823 | -0.828 | -0.5836 | Yes | ||
| 59 | POLS | 5890438 | 17840 | -0.842 | -0.5628 | Yes | ||
| 60 | DYRK1A | 3190181 | 18053 | -1.036 | -0.5475 | Yes | ||
| 61 | SOX4 | 2260091 | 18158 | -1.199 | -0.5223 | Yes | ||
| 62 | CTCF | 5340017 | 18177 | -1.215 | -0.4919 | Yes | ||
| 63 | CRKL | 4050427 | 18189 | -1.245 | -0.4604 | Yes | ||
| 64 | WSB1 | 870563 | 18195 | -1.252 | -0.4285 | Yes | ||
| 65 | MARK3 | 2370450 | 18196 | -1.253 | -0.3962 | Yes | ||
| 66 | BRD1 | 1660278 | 18199 | -1.258 | -0.3639 | Yes | ||
| 67 | SPOP | 450035 | 18217 | -1.298 | -0.3314 | Yes | ||
| 68 | IL4R | 2370520 | 18245 | -1.357 | -0.2979 | Yes | ||
| 69 | NEDD4L | 6380368 | 18298 | -1.475 | -0.2627 | Yes | ||
| 70 | KLF7 | 50390 | 18299 | -1.477 | -0.2247 | Yes | ||
| 71 | KBTBD2 | 2320014 | 18334 | -1.590 | -0.1855 | Yes | ||
| 72 | ARL4C | 1190279 | 18377 | -1.720 | -0.1435 | Yes | ||
| 73 | TNFAIP3 | 2900142 | 18460 | -1.987 | -0.0967 | Yes | ||
| 74 | TNFRSF1A | 1090390 6520735 | 18609 | -4.080 | 0.0004 | Yes |

