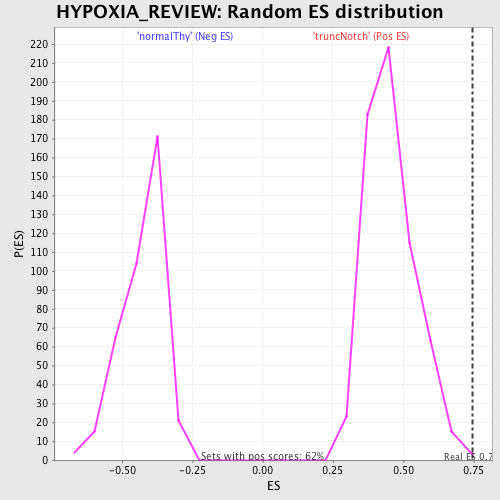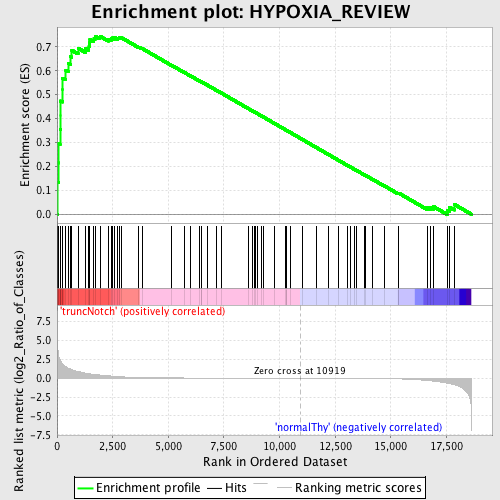
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | truncNotch |
| GeneSet | HYPOXIA_REVIEW |
| Enrichment Score (ES) | 0.74497163 |
| Normalized Enrichment Score (NES) | 1.6415937 |
| Nominal p-value | 0.0048387097 |
| FDR q-value | 0.040579338 |
| FWER p-Value | 0.347 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PGK1 | 1570494 630300 | 11 | 5.024 | 0.1346 | Yes | ||
| 2 | HMOX1 | 1740687 | 64 | 3.070 | 0.2144 | Yes | ||
| 3 | APEX1 | 3190519 | 70 | 2.994 | 0.2947 | Yes | ||
| 4 | IGF2 | 6510020 | 134 | 2.394 | 0.3557 | Yes | ||
| 5 | PKM2 | 6520403 70500 | 160 | 2.217 | 0.4140 | Yes | ||
| 6 | ENO1 | 5340128 | 161 | 2.215 | 0.4736 | Yes | ||
| 7 | ALDOA | 6290672 | 242 | 1.875 | 0.5197 | Yes | ||
| 8 | CDKN1A | 4050088 6400706 | 246 | 1.863 | 0.5697 | Yes | ||
| 9 | MIF | 3710332 | 391 | 1.500 | 0.6023 | Yes | ||
| 10 | TGM2 | 5360452 | 502 | 1.326 | 0.6321 | Yes | ||
| 11 | TFRC | 4050551 | 596 | 1.200 | 0.6593 | Yes | ||
| 12 | PFKL | 6200167 | 661 | 1.110 | 0.6858 | Yes | ||
| 13 | LDHA | 2190594 | 944 | 0.881 | 0.6942 | Yes | ||
| 14 | HK2 | 2640722 | 1283 | 0.665 | 0.6939 | Yes | ||
| 15 | SLC2A3 | 1990377 | 1432 | 0.592 | 0.7019 | Yes | ||
| 16 | SPP1 | 2470609 | 1451 | 0.584 | 0.7166 | Yes | ||
| 17 | COL5A1 | 2230050 | 1474 | 0.572 | 0.7308 | Yes | ||
| 18 | ADM | 4810121 | 1657 | 0.502 | 0.7345 | Yes | ||
| 19 | TGFB1 | 1940162 | 1706 | 0.484 | 0.7450 | Yes | ||
| 20 | NFKB1 | 5420358 | 1928 | 0.422 | 0.7444 | No | ||
| 21 | BHLHB2 | 7040603 | 2302 | 0.317 | 0.7328 | No | ||
| 22 | PLAUR | 5910280 | 2429 | 0.284 | 0.7337 | No | ||
| 23 | TFF3 | 1580129 | 2494 | 0.265 | 0.7374 | No | ||
| 24 | CDKN1B | 3800025 6450044 | 2587 | 0.243 | 0.7389 | No | ||
| 25 | SLC2A1 | 2100609 | 2733 | 0.208 | 0.7367 | No | ||
| 26 | TGFB3 | 1070041 | 2790 | 0.197 | 0.7390 | No | ||
| 27 | VIM | 20431 | 2895 | 0.181 | 0.7383 | No | ||
| 28 | EPAS1 | 5290156 | 3671 | 0.095 | 0.6991 | No | ||
| 29 | PGF | 4810593 | 3817 | 0.086 | 0.6936 | No | ||
| 30 | FLT1 | 3830167 4920438 | 5147 | 0.038 | 0.6229 | No | ||
| 31 | HK1 | 4280402 | 5714 | 0.028 | 0.5932 | No | ||
| 32 | STC1 | 360161 | 5992 | 0.024 | 0.5789 | No | ||
| 33 | EPO | 940180 | 6410 | 0.019 | 0.5569 | No | ||
| 34 | GAPDH | 110022 430039 6220161 | 6478 | 0.019 | 0.5538 | No | ||
| 35 | CCL2 | 4760019 | 6494 | 0.019 | 0.5535 | No | ||
| 36 | FGF3 | 4230685 | 6737 | 0.017 | 0.5409 | No | ||
| 37 | CP | 2570484 | 7148 | 0.014 | 0.5191 | No | ||
| 38 | FOS | 1850315 | 7390 | 0.012 | 0.5065 | No | ||
| 39 | TEK | 6350541 | 8601 | 0.007 | 0.4414 | No | ||
| 40 | TH | 2100056 | 8781 | 0.006 | 0.4319 | No | ||
| 41 | ENPEP | 130373 | 8849 | 0.006 | 0.4285 | No | ||
| 42 | ANGPT2 | 1230458 5720706 6040605 | 8928 | 0.006 | 0.4244 | No | ||
| 43 | PTGS2 | 2510301 3170369 | 9010 | 0.005 | 0.4202 | No | ||
| 44 | MMP13 | 510471 3940097 | 9175 | 0.005 | 0.4115 | No | ||
| 45 | EDN2 | 6760647 | 9269 | 0.005 | 0.4066 | No | ||
| 46 | HIF1A | 5670605 | 9787 | 0.003 | 0.3788 | No | ||
| 47 | TAGLN | 6660280 | 10269 | 0.002 | 0.3529 | No | ||
| 48 | XRCC5 | 7100286 | 10319 | 0.002 | 0.3504 | No | ||
| 49 | BNIP3 | 3140270 | 10327 | 0.002 | 0.3500 | No | ||
| 50 | IGFBP1 | 5720435 | 10500 | 0.001 | 0.3408 | No | ||
| 51 | IGFBP3 | 2370500 | 11032 | -0.000 | 0.3122 | No | ||
| 52 | HGF | 3360593 | 11642 | -0.002 | 0.2794 | No | ||
| 53 | EDN1 | 1770047 | 12188 | -0.004 | 0.2501 | No | ||
| 54 | F3 | 2940180 | 12655 | -0.006 | 0.2252 | No | ||
| 55 | HDAC9 | 1990010 2260133 | 13034 | -0.008 | 0.2050 | No | ||
| 56 | TGFA | 1570332 | 13191 | -0.009 | 0.1968 | No | ||
| 57 | IGFBP2 | 5220563 | 13387 | -0.011 | 0.1866 | No | ||
| 58 | IL6 | 380133 | 13456 | -0.012 | 0.1833 | No | ||
| 59 | L1CAM | 4850021 | 13818 | -0.016 | 0.1642 | No | ||
| 60 | JUN | 840170 | 13846 | -0.016 | 0.1632 | No | ||
| 61 | LRP8 | 3610746 5360035 | 14160 | -0.022 | 0.1469 | No | ||
| 62 | PRPS1 | 6350129 | 14698 | -0.040 | 0.1191 | No | ||
| 63 | PDGFB | 3060440 6370008 | 15328 | -0.083 | 0.0874 | No | ||
| 64 | P4HA1 | 2630692 | 15364 | -0.087 | 0.0878 | No | ||
| 65 | BNIP3L | 1940347 | 16639 | -0.303 | 0.0273 | No | ||
| 66 | DDIT3 | 780373 | 16773 | -0.341 | 0.0293 | No | ||
| 67 | SAT1 | 4570463 | 16922 | -0.387 | 0.0318 | No | ||
| 68 | PFKP | 70138 6760040 1170278 | 17534 | -0.650 | 0.0163 | No | ||
| 69 | CCNG2 | 3190095 | 17629 | -0.698 | 0.0300 | No | ||
| 70 | XRCC6 | 2850450 3870528 | 17862 | -0.861 | 0.0407 | No |