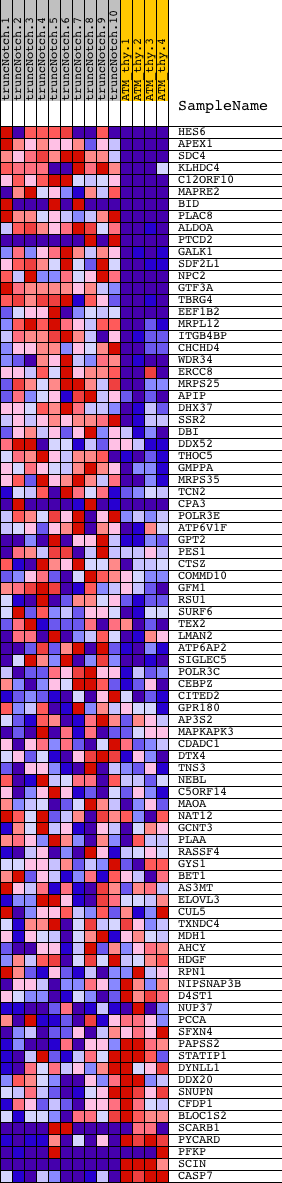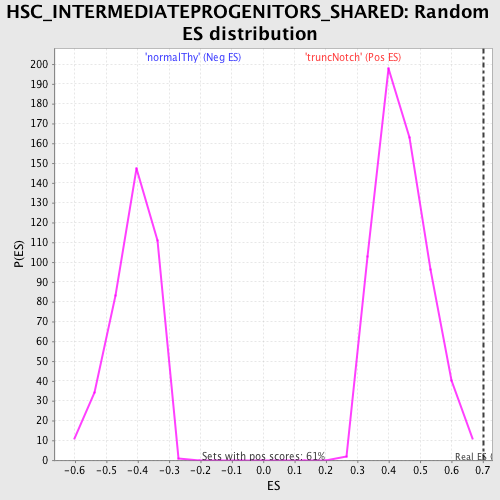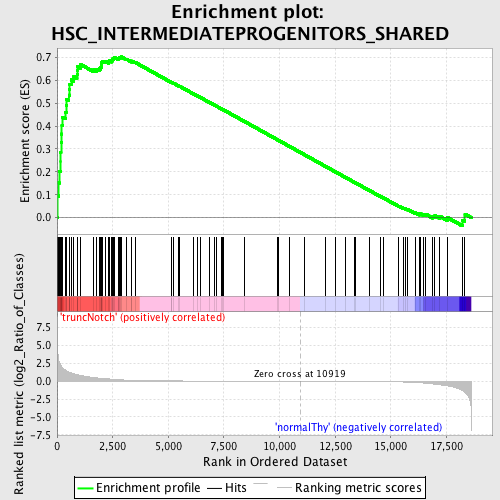
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | truncNotch |
| GeneSet | HSC_INTERMEDIATEPROGENITORS_SHARED |
| Enrichment Score (ES) | 0.7022185 |
| Normalized Enrichment Score (NES) | 1.5854497 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.09889006 |
| FWER p-Value | 0.812 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HES6 | 540411 6550504 | 13 | 4.961 | 0.0967 | Yes | ||
| 2 | APEX1 | 3190519 | 70 | 2.994 | 0.1524 | Yes | ||
| 3 | SDC4 | 6370411 | 94 | 2.683 | 0.2039 | Yes | ||
| 4 | KLHDC4 | 870288 5900092 | 163 | 2.207 | 0.2435 | Yes | ||
| 5 | C12ORF10 | 4120324 5270113 | 165 | 2.202 | 0.2867 | Yes | ||
| 6 | MAPRE2 | 1660435 5860403 | 175 | 2.141 | 0.3283 | Yes | ||
| 7 | BID | 4590592 6130008 | 208 | 1.985 | 0.3655 | Yes | ||
| 8 | PLAC8 | 2900019 | 217 | 1.948 | 0.4033 | Yes | ||
| 9 | ALDOA | 6290672 | 242 | 1.875 | 0.4388 | Yes | ||
| 10 | PTCD2 | 1230088 1400605 3170576 6200609 | 385 | 1.522 | 0.4610 | Yes | ||
| 11 | GALK1 | 840162 | 408 | 1.475 | 0.4888 | Yes | ||
| 12 | SDF2L1 | 4230446 | 425 | 1.453 | 0.5165 | Yes | ||
| 13 | NPC2 | 2350102 | 534 | 1.280 | 0.5357 | Yes | ||
| 14 | GTF3A | 2340451 | 555 | 1.261 | 0.5594 | Yes | ||
| 15 | TBRG4 | 3610239 6370594 | 570 | 1.247 | 0.5831 | Yes | ||
| 16 | EEF1B2 | 4850400 6130161 | 638 | 1.140 | 0.6019 | Yes | ||
| 17 | MRPL12 | 3120112 | 750 | 1.030 | 0.6161 | Yes | ||
| 18 | ITGB4BP | 1400541 | 894 | 0.919 | 0.6265 | Yes | ||
| 19 | CHCHD4 | 2680075 | 912 | 0.905 | 0.6433 | Yes | ||
| 20 | WDR34 | 6100215 | 925 | 0.894 | 0.6602 | Yes | ||
| 21 | ERCC8 | 1240300 6450372 6590180 | 1063 | 0.804 | 0.6686 | Yes | ||
| 22 | MRPS25 | 610441 870128 1410368 | 1650 | 0.505 | 0.6469 | Yes | ||
| 23 | APIP | 2480440 2810471 | 1790 | 0.458 | 0.6484 | Yes | ||
| 24 | DHX37 | 1400309 | 1898 | 0.429 | 0.6511 | Yes | ||
| 25 | SSR2 | 5390180 | 1959 | 0.411 | 0.6559 | Yes | ||
| 26 | DBI | 1090706 | 1988 | 0.399 | 0.6622 | Yes | ||
| 27 | DDX52 | 2970411 | 1990 | 0.398 | 0.6700 | Yes | ||
| 28 | THOC5 | 4570047 | 2002 | 0.393 | 0.6771 | Yes | ||
| 29 | GMPPA | 520450 3120497 | 2026 | 0.388 | 0.6835 | Yes | ||
| 30 | MRPS35 | 2370603 4070551 | 2172 | 0.351 | 0.6825 | Yes | ||
| 31 | TCN2 | 2810139 | 2323 | 0.312 | 0.6806 | Yes | ||
| 32 | CPA3 | 610242 6380102 | 2333 | 0.309 | 0.6861 | Yes | ||
| 33 | POLR3E | 380152 | 2456 | 0.279 | 0.6850 | Yes | ||
| 34 | ATP6V1F | 5420082 | 2476 | 0.271 | 0.6893 | Yes | ||
| 35 | GPT2 | 1850463 | 2505 | 0.263 | 0.6930 | Yes | ||
| 36 | PES1 | 2470427 | 2548 | 0.253 | 0.6957 | Yes | ||
| 37 | CTSZ | 1500687 1690364 | 2578 | 0.245 | 0.6989 | Yes | ||
| 38 | COMMD10 | 3120451 | 2764 | 0.202 | 0.6929 | Yes | ||
| 39 | GFM1 | 6220711 | 2767 | 0.201 | 0.6967 | Yes | ||
| 40 | RSU1 | 4850372 | 2788 | 0.198 | 0.6995 | Yes | ||
| 41 | SURF6 | 3060451 | 2840 | 0.190 | 0.7005 | Yes | ||
| 42 | TEX2 | 1740504 2480102 3060494 | 2876 | 0.184 | 0.7022 | Yes | ||
| 43 | LMAN2 | 430154 | 3122 | 0.149 | 0.6919 | No | ||
| 44 | ATP6AP2 | 4010059 5080315 7100347 6660484 | 3353 | 0.121 | 0.6819 | No | ||
| 45 | SIGLEC5 | 5690398 | 3355 | 0.121 | 0.6842 | No | ||
| 46 | POLR3C | 6940546 | 3509 | 0.108 | 0.6781 | No | ||
| 47 | CEBPZ | 2970053 2680068 | 5157 | 0.038 | 0.5899 | No | ||
| 48 | CITED2 | 5670114 5130088 | 5161 | 0.038 | 0.5905 | No | ||
| 49 | GPR180 | 5390711 | 5249 | 0.036 | 0.5865 | No | ||
| 50 | AP3S2 | 2630411 5080053 | 5468 | 0.032 | 0.5754 | No | ||
| 51 | MAPKAPK3 | 630520 | 5500 | 0.031 | 0.5743 | No | ||
| 52 | CDADC1 | 3710201 4120064 | 6115 | 0.022 | 0.5416 | No | ||
| 53 | DTX4 | 4120433 | 6288 | 0.020 | 0.5327 | No | ||
| 54 | TNS3 | 6380242 | 6425 | 0.019 | 0.5258 | No | ||
| 55 | NEBL | 1850369 2480286 | 6833 | 0.016 | 0.5041 | No | ||
| 56 | C5ORF14 | 3780050 | 7078 | 0.014 | 0.4912 | No | ||
| 57 | MAOA | 1410039 4610324 | 7153 | 0.014 | 0.4875 | No | ||
| 58 | NAT12 | 1780092 4730370 | 7374 | 0.012 | 0.4759 | No | ||
| 59 | GCNT3 | 6290239 | 7449 | 0.012 | 0.4721 | No | ||
| 60 | PLAA | 60706 6370369 | 7467 | 0.012 | 0.4714 | No | ||
| 61 | RASSF4 | 510372 1660673 6290040 | 8434 | 0.008 | 0.4194 | No | ||
| 62 | GYS1 | 540154 | 9913 | 0.003 | 0.3397 | No | ||
| 63 | BET1 | 4010725 | 9934 | 0.003 | 0.3387 | No | ||
| 64 | AS3MT | 1570064 2350113 | 10438 | 0.001 | 0.3116 | No | ||
| 65 | ELOVL3 | 6860050 | 11133 | -0.001 | 0.2741 | No | ||
| 66 | CUL5 | 450142 | 12053 | -0.004 | 0.2246 | No | ||
| 67 | TXNDC4 | 450537 | 12515 | -0.006 | 0.1998 | No | ||
| 68 | MDH1 | 6660358 6760731 | 12968 | -0.008 | 0.1756 | No | ||
| 69 | AHCY | 4230605 | 13382 | -0.011 | 0.1535 | No | ||
| 70 | HDGF | 5130114 5420066 6590152 | 13419 | -0.011 | 0.1518 | No | ||
| 71 | RPN1 | 1240113 | 14045 | -0.020 | 0.1185 | No | ||
| 72 | NIPSNAP3B | 630672 3710091 4540288 4780576 | 14537 | -0.033 | 0.0926 | No | ||
| 73 | D4ST1 | 1780671 | 14686 | -0.040 | 0.0854 | No | ||
| 74 | NUP37 | 2370097 6370435 6380008 | 15360 | -0.086 | 0.0508 | No | ||
| 75 | PCCA | 3390400 | 15586 | -0.110 | 0.0408 | No | ||
| 76 | SFXN4 | 60750 4560059 | 15650 | -0.117 | 0.0397 | No | ||
| 77 | PAPSS2 | 870113 | 15764 | -0.130 | 0.0361 | No | ||
| 78 | STATIP1 | 4480022 6940097 | 16090 | -0.176 | 0.0220 | No | ||
| 79 | DYNLL1 | 5570113 | 16284 | -0.213 | 0.0158 | No | ||
| 80 | DDX20 | 3290348 | 16325 | -0.221 | 0.0180 | No | ||
| 81 | SNUPN | 5050131 | 16455 | -0.251 | 0.0160 | No | ||
| 82 | CFDP1 | 7000112 | 16562 | -0.280 | 0.0157 | No | ||
| 83 | BLOC1S2 | 1240215 | 16884 | -0.374 | 0.0057 | No | ||
| 84 | SCARB1 | 510441 6770047 | 16981 | -0.408 | 0.0086 | No | ||
| 85 | PYCARD | 2100750 | 17196 | -0.490 | 0.0066 | No | ||
| 86 | PFKP | 70138 6760040 1170278 | 17534 | -0.650 | 0.0012 | No | ||
| 87 | SCIN | 5420180 | 18226 | -1.318 | -0.0102 | No | ||
| 88 | CASP7 | 4210156 | 18333 | -1.590 | 0.0153 | No |