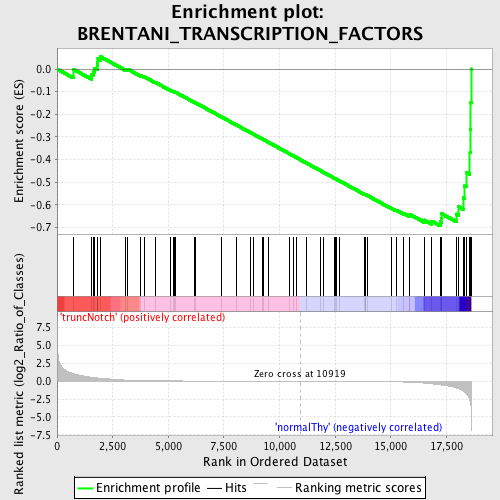
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | BRENTANI_TRANSCRIPTION_FACTORS |
| Enrichment Score (ES) | -0.6930934 |
| Normalized Enrichment Score (NES) | -1.5722666 |
| Nominal p-value | 0.0025974025 |
| FDR q-value | 0.30623907 |
| FWER p-Value | 0.969 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | STAT5A | 2680458 | 724 | 1.049 | -0.0007 | No | ||
| 2 | AATF | 5220575 | 1534 | 0.548 | -0.0242 | No | ||
| 3 | SPI1 | 1410397 | 1639 | 0.511 | -0.0111 | No | ||
| 4 | MYBL2 | 5220609 6520368 | 1685 | 0.492 | 0.0045 | No | ||
| 5 | NME1 | 770014 | 1804 | 0.453 | 0.0147 | No | ||
| 6 | MAFG | 4120440 | 1814 | 0.450 | 0.0307 | No | ||
| 7 | STAT6 | 1190010 5720019 | 1834 | 0.444 | 0.0459 | No | ||
| 8 | LITAF | 6940671 | 1964 | 0.410 | 0.0539 | No | ||
| 9 | EGR3 | 6940128 | 3087 | 0.153 | -0.0009 | No | ||
| 10 | ETS1 | 5270278 6450717 6620465 | 3175 | 0.142 | -0.0004 | No | ||
| 11 | ETV6 | 610524 | 3767 | 0.089 | -0.0290 | No | ||
| 12 | PML | 50093 2190435 2450402 3840082 | 3908 | 0.080 | -0.0336 | No | ||
| 13 | WT1 | 3990463 | 4400 | 0.058 | -0.0579 | No | ||
| 14 | GLI2 | 3060632 | 5094 | 0.039 | -0.0938 | No | ||
| 15 | FOSL2 | 870044 1230427 6520433 | 5215 | 0.037 | -0.0989 | No | ||
| 16 | GLI3 | 5690148 | 5282 | 0.035 | -0.1012 | No | ||
| 17 | CEBPA | 5690519 | 5330 | 0.035 | -0.1025 | No | ||
| 18 | ETV1 | 70735 2940603 5080463 | 6177 | 0.022 | -0.1473 | No | ||
| 19 | ATF1 | 4570142 5550095 | 6230 | 0.021 | -0.1493 | No | ||
| 20 | FOS | 1850315 | 7390 | 0.012 | -0.2113 | No | ||
| 21 | AHR | 4560671 | 8072 | 0.009 | -0.2477 | No | ||
| 22 | REL | 360707 | 8673 | 0.007 | -0.2798 | No | ||
| 23 | DPF2 | 6980373 | 8840 | 0.006 | -0.2885 | No | ||
| 24 | PBX1 | 6660301 | 9232 | 0.005 | -0.3094 | No | ||
| 25 | TLX1 | 2630162 | 9298 | 0.005 | -0.3127 | No | ||
| 26 | CHES1 | 4570121 5700162 | 9508 | 0.004 | -0.3238 | No | ||
| 27 | EHF | 4560088 | 10424 | 0.001 | -0.3731 | No | ||
| 28 | LMO1 | 3170528 | 10605 | 0.001 | -0.3828 | No | ||
| 29 | MYC | 380541 4670170 | 10764 | 0.000 | -0.3913 | No | ||
| 30 | MLLT7 | 4480707 | 11196 | -0.001 | -0.4144 | No | ||
| 31 | FOSL1 | 430021 | 11830 | -0.003 | -0.4485 | No | ||
| 32 | MYOD1 | 1230731 | 11994 | -0.003 | -0.4571 | No | ||
| 33 | RXRA | 3800471 | 12485 | -0.005 | -0.4833 | No | ||
| 34 | PPARA | 2060026 | 12517 | -0.006 | -0.4848 | No | ||
| 35 | FOXD1 | 6860053 | 12535 | -0.006 | -0.4855 | No | ||
| 36 | PAX7 | 6860593 | 12693 | -0.006 | -0.4937 | No | ||
| 37 | MYBL1 | 2900168 | 13835 | -0.016 | -0.5546 | No | ||
| 38 | JUN | 840170 | 13846 | -0.016 | -0.5546 | No | ||
| 39 | FOSB | 1940142 | 13859 | -0.016 | -0.5546 | No | ||
| 40 | STAT5B | 6200026 | 13947 | -0.018 | -0.5586 | No | ||
| 41 | FOXD2 | 4210035 | 15033 | -0.060 | -0.6149 | No | ||
| 42 | RELB | 1400048 | 15248 | -0.075 | -0.6237 | No | ||
| 43 | NFKB2 | 2320670 | 15588 | -0.110 | -0.6379 | No | ||
| 44 | TCF7L2 | 2190348 5220113 | 15855 | -0.142 | -0.6471 | No | ||
| 45 | ETV3 | 5360487 | 15857 | -0.142 | -0.6419 | No | ||
| 46 | IRF4 | 610133 | 16497 | -0.264 | -0.6667 | No | ||
| 47 | RELA | 3830075 | 16834 | -0.358 | -0.6717 | No | ||
| 48 | FLI1 | 3990142 | 17232 | -0.505 | -0.6746 | Yes | ||
| 49 | MYB | 1660494 5860451 6130706 | 17283 | -0.524 | -0.6582 | Yes | ||
| 50 | JUNB | 4230048 | 17292 | -0.526 | -0.6394 | Yes | ||
| 51 | SMARCA4 | 6980528 | 17944 | -0.936 | -0.6402 | Yes | ||
| 52 | STAT3 | 460040 3710341 | 18039 | -1.029 | -0.6076 | Yes | ||
| 53 | RUNX1 | 3840711 | 18253 | -1.370 | -0.5690 | Yes | ||
| 54 | CBFB | 1850551 2650035 | 18316 | -1.528 | -0.5164 | Yes | ||
| 55 | PBX2 | 3130433 6510292 | 18400 | -1.770 | -0.4561 | Yes | ||
| 56 | EGR1 | 4610347 | 18557 | -2.630 | -0.3683 | Yes | ||
| 57 | ETS2 | 360451 | 18573 | -2.835 | -0.2654 | Yes | ||
| 58 | SRF | 70139 2320039 | 18598 | -3.207 | -0.1493 | Yes | ||
| 59 | BCL6 | 940100 | 18610 | -4.108 | 0.0003 | Yes |

