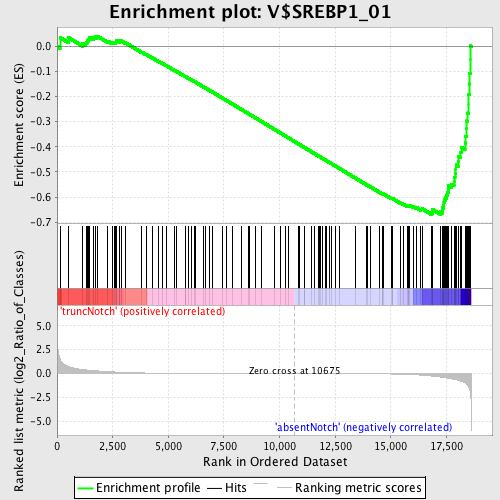
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #truncNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#truncNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | V$SREBP1_01 |
| Enrichment Score (ES) | -0.66955024 |
| Normalized Enrichment Score (NES) | -1.7066163 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0074335556 |
| FWER p-Value | 0.047 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATP6V0D1 | 360348 5900021 6290605 | 131 | 1.525 | 0.0353 | No | ||
| 2 | BLOC1S1 | 130341 3940717 | 522 | 0.724 | 0.0343 | No | ||
| 3 | BLNK | 70288 6590092 | 1154 | 0.414 | 0.0117 | No | ||
| 4 | SDF2 | 2690601 | 1298 | 0.375 | 0.0143 | No | ||
| 5 | BAX | 3830008 | 1345 | 0.365 | 0.0220 | No | ||
| 6 | LCP1 | 4280397 | 1393 | 0.353 | 0.0292 | No | ||
| 7 | MRPL27 | 2060131 | 1462 | 0.337 | 0.0349 | No | ||
| 8 | CAMK2D | 2370685 | 1639 | 0.305 | 0.0339 | No | ||
| 9 | ANAPC13 | 3800446 | 1710 | 0.292 | 0.0382 | No | ||
| 10 | HTATIP | 5360739 | 1805 | 0.276 | 0.0408 | No | ||
| 11 | CBARA1 | 360471 | 2283 | 0.204 | 0.0206 | No | ||
| 12 | ALG1 | 2570112 | 2484 | 0.175 | 0.0147 | No | ||
| 13 | RAB3A | 3940288 | 2565 | 0.166 | 0.0150 | No | ||
| 14 | SNRPA | 2570100 4150121 6940484 | 2639 | 0.156 | 0.0154 | No | ||
| 15 | ASNA1 | 3830041 | 2647 | 0.154 | 0.0193 | No | ||
| 16 | PIAS4 | 6040113 | 2662 | 0.151 | 0.0227 | No | ||
| 17 | EPC1 | 1570050 5290095 6900193 | 2790 | 0.135 | 0.0196 | No | ||
| 18 | GNB2 | 2350053 | 2818 | 0.132 | 0.0218 | No | ||
| 19 | ASPSCR1 | 610167 3990739 | 2885 | 0.126 | 0.0217 | No | ||
| 20 | CLN3 | 780368 | 3067 | 0.108 | 0.0149 | No | ||
| 21 | HOXB7 | 4540706 4610358 | 3802 | 0.063 | -0.0230 | No | ||
| 22 | TOP3A | 7000435 | 4006 | 0.056 | -0.0324 | No | ||
| 23 | POLR3C | 6940546 | 4275 | 0.048 | -0.0456 | No | ||
| 24 | GTF2H1 | 2570746 2650148 | 4577 | 0.040 | -0.0608 | No | ||
| 25 | HIVEP1 | 3850632 | 4737 | 0.037 | -0.0683 | No | ||
| 26 | SNAI1 | 6590253 | 4903 | 0.034 | -0.0763 | No | ||
| 27 | DUSP8 | 6650039 | 5272 | 0.027 | -0.0955 | No | ||
| 28 | MYST2 | 4540494 | 5386 | 0.025 | -0.1009 | No | ||
| 29 | DLX2 | 2320438 4200673 | 5762 | 0.021 | -0.1206 | No | ||
| 30 | GGN | 1410528 4210082 5290170 | 5777 | 0.021 | -0.1207 | No | ||
| 31 | MCM2 | 5050139 | 5911 | 0.020 | -0.1274 | No | ||
| 32 | RNF146 | 1660735 | 6047 | 0.018 | -0.1342 | No | ||
| 33 | USH1G | 2940446 | 6179 | 0.017 | -0.1408 | No | ||
| 34 | HOXB5 | 2900538 | 6224 | 0.017 | -0.1427 | No | ||
| 35 | OTOP2 | 610068 | 6559 | 0.014 | -0.1603 | No | ||
| 36 | ADCY2 | 4610100 | 6662 | 0.014 | -0.1655 | No | ||
| 37 | RAD9A | 110300 1940632 3990390 6040014 | 6843 | 0.013 | -0.1748 | No | ||
| 38 | RGL1 | 1500097 6900450 | 6979 | 0.012 | -0.1818 | No | ||
| 39 | IRS4 | 430341 1190450 | 7432 | 0.010 | -0.2060 | No | ||
| 40 | SEPT3 | 3830022 5290079 | 7624 | 0.009 | -0.2161 | No | ||
| 41 | HOXC5 | 1580017 | 7873 | 0.008 | -0.2293 | No | ||
| 42 | SLC35A5 | 2970670 4480546 | 8287 | 0.007 | -0.2514 | No | ||
| 43 | FMR1 | 5050075 | 8609 | 0.006 | -0.2686 | No | ||
| 44 | BDNF | 2940128 3520368 | 8662 | 0.005 | -0.2713 | No | ||
| 45 | SLC36A1 | 5080242 | 8909 | 0.005 | -0.2844 | No | ||
| 46 | DVL2 | 6110162 | 9180 | 0.004 | -0.2989 | No | ||
| 47 | ATP7A | 6550168 | 9778 | 0.002 | -0.3312 | No | ||
| 48 | SOX10 | 6200538 | 10051 | 0.002 | -0.3458 | No | ||
| 49 | HOXA11 | 6980133 | 10256 | 0.001 | -0.3568 | No | ||
| 50 | SMCR8 | 730653 | 10404 | 0.001 | -0.3647 | No | ||
| 51 | TCEAL1 | 870577 | 10870 | -0.000 | -0.3899 | No | ||
| 52 | ABR | 610079 1170609 3610195 5670050 | 10895 | -0.001 | -0.3912 | No | ||
| 53 | KIAA1715 | 1190551 | 11110 | -0.001 | -0.4027 | No | ||
| 54 | XPO1 | 540707 | 11119 | -0.001 | -0.4031 | No | ||
| 55 | KLF12 | 1660095 4810288 5340546 6520286 | 11455 | -0.002 | -0.4212 | No | ||
| 56 | UBE2B | 4780497 | 11557 | -0.002 | -0.4266 | No | ||
| 57 | CUL5 | 450142 | 11747 | -0.003 | -0.4367 | No | ||
| 58 | TRPM7 | 2510408 | 11802 | -0.003 | -0.4395 | No | ||
| 59 | ACCN1 | 2060139 2190541 | 11824 | -0.003 | -0.4406 | No | ||
| 60 | PIP5K1A | 4560672 | 11918 | -0.003 | -0.4455 | No | ||
| 61 | RRM2B | 1690025 1690465 4010400 | 12074 | -0.004 | -0.4538 | No | ||
| 62 | PRKCE | 5700053 | 12122 | -0.004 | -0.4562 | No | ||
| 63 | SET | 6650286 | 12246 | -0.004 | -0.4627 | No | ||
| 64 | ARPC5 | 1740411 3450300 | 12353 | -0.005 | -0.4683 | No | ||
| 65 | GLA | 4610364 | 12532 | -0.006 | -0.4778 | No | ||
| 66 | HNRPH2 | 2630040 5720671 | 12706 | -0.006 | -0.4870 | No | ||
| 67 | HOXA2 | 2120121 | 13430 | -0.010 | -0.5258 | No | ||
| 68 | ZIC3 | 380020 | 13919 | -0.015 | -0.5518 | No | ||
| 69 | CANX | 4760402 | 13932 | -0.015 | -0.5520 | No | ||
| 70 | HOXA7 | 5910152 | 14071 | -0.017 | -0.5590 | No | ||
| 71 | ARMCX3 | 3290309 | 14511 | -0.025 | -0.5820 | No | ||
| 72 | DSCAM | 1780050 2450731 2810438 | 14622 | -0.029 | -0.5872 | No | ||
| 73 | FEN1 | 1770541 | 14640 | -0.029 | -0.5873 | No | ||
| 74 | DIRC2 | 2810494 | 14667 | -0.031 | -0.5878 | No | ||
| 75 | APEX1 | 3190519 | 15039 | -0.045 | -0.6066 | No | ||
| 76 | SNX16 | 870446 | 15055 | -0.046 | -0.6062 | No | ||
| 77 | PICALM | 1940280 | 15058 | -0.046 | -0.6050 | No | ||
| 78 | MTCH2 | 3780603 6620215 | 15440 | -0.068 | -0.6237 | No | ||
| 79 | QTRT1 | 1340162 | 15586 | -0.078 | -0.6293 | No | ||
| 80 | CHD2 | 4070039 | 15749 | -0.092 | -0.6356 | No | ||
| 81 | OSGEP | 5700239 | 15794 | -0.096 | -0.6353 | No | ||
| 82 | POLDIP2 | 2650139 | 15808 | -0.097 | -0.6333 | No | ||
| 83 | TNRC15 | 6760746 | 15849 | -0.100 | -0.6327 | No | ||
| 84 | ZFYVE26 | 6860152 | 16007 | -0.118 | -0.6379 | No | ||
| 85 | RAB22A | 110500 3830707 | 16144 | -0.130 | -0.6416 | No | ||
| 86 | HSPBAP1 | 4060022 | 16343 | -0.160 | -0.6479 | No | ||
| 87 | PDCD6IP | 1990403 | 16409 | -0.171 | -0.6467 | No | ||
| 88 | SIRT1 | 1190731 | 16815 | -0.257 | -0.6614 | Yes | ||
| 89 | DAZAP2 | 6200102 | 16868 | -0.269 | -0.6568 | Yes | ||
| 90 | SOCS2 | 4760692 | 16870 | -0.269 | -0.6494 | Yes | ||
| 91 | EIF4B | 5390563 5270577 | 17244 | -0.369 | -0.6593 | Yes | ||
| 92 | LAMP1 | 2470524 | 17329 | -0.395 | -0.6529 | Yes | ||
| 93 | STX6 | 4850750 6770278 | 17338 | -0.398 | -0.6423 | Yes | ||
| 94 | SUPT16H | 1240333 | 17377 | -0.413 | -0.6328 | Yes | ||
| 95 | ATP6V1A | 6590242 | 17388 | -0.416 | -0.6218 | Yes | ||
| 96 | GRN | 4050239 | 17410 | -0.422 | -0.6113 | Yes | ||
| 97 | GSK3B | 5360348 | 17444 | -0.435 | -0.6010 | Yes | ||
| 98 | NGFRAP1 | 6770195 | 17512 | -0.459 | -0.5918 | Yes | ||
| 99 | EIF4G1 | 4070446 | 17543 | -0.470 | -0.5804 | Yes | ||
| 100 | HPS3 | 6650348 | 17581 | -0.489 | -0.5688 | Yes | ||
| 101 | AARS | 1570279 2370136 | 17592 | -0.493 | -0.5557 | Yes | ||
| 102 | STMN1 | 1990717 | 17737 | -0.551 | -0.5482 | Yes | ||
| 103 | ALDH6A1 | 1770746 | 17842 | -0.598 | -0.5372 | Yes | ||
| 104 | IGF2R | 1570402 | 17847 | -0.602 | -0.5207 | Yes | ||
| 105 | COMMD3 | 2510259 | 17886 | -0.619 | -0.5056 | Yes | ||
| 106 | DCTN4 | 3390010 5550170 | 17926 | -0.639 | -0.4900 | Yes | ||
| 107 | TOPORS | 4060592 | 17929 | -0.640 | -0.4723 | Yes | ||
| 108 | HK2 | 2640722 | 18028 | -0.700 | -0.4582 | Yes | ||
| 109 | RRAGC | 5910348 | 18037 | -0.703 | -0.4391 | Yes | ||
| 110 | CCAR1 | 3190092 | 18130 | -0.771 | -0.4226 | Yes | ||
| 111 | SUPT6H | 3170082 | 18163 | -0.801 | -0.4021 | Yes | ||
| 112 | ZDHHC13 | 430170 4230021 | 18336 | -0.985 | -0.3841 | Yes | ||
| 113 | NR1D1 | 2360471 770746 6590204 | 18363 | -1.024 | -0.3570 | Yes | ||
| 114 | AKAP12 | 1450739 | 18414 | -1.125 | -0.3285 | Yes | ||
| 115 | CSRP2BP | 4570717 5550333 | 18420 | -1.140 | -0.2971 | Yes | ||
| 116 | SNX2 | 2470068 3360364 6510064 | 18432 | -1.165 | -0.2654 | Yes | ||
| 117 | GPNMB | 1780138 2340176 3390403 4560156 | 18503 | -1.401 | -0.2303 | Yes | ||
| 118 | CD164 | 3830594 | 18504 | -1.406 | -0.1912 | Yes | ||
| 119 | ATF7IP | 2690176 6770021 | 18529 | -1.539 | -0.1498 | Yes | ||
| 120 | ATF4 | 730441 1740195 | 18536 | -1.582 | -0.1062 | Yes | ||
| 121 | U2AF2 | 450600 4670128 5420292 | 18568 | -1.982 | -0.0529 | Yes | ||
| 122 | UBE4B | 780008 3610154 | 18570 | -1.996 | 0.0025 | Yes |

