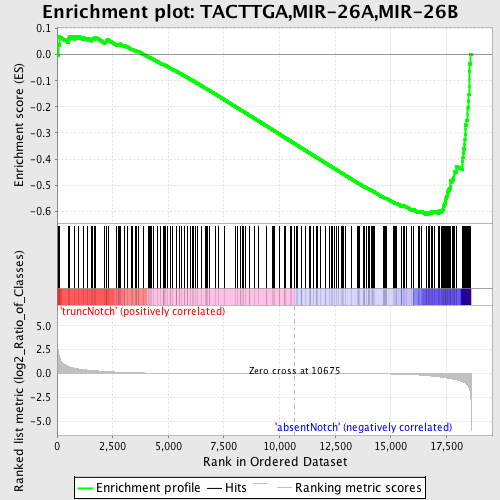
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #truncNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#truncNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | TACTTGA,MIR-26A,MIR-26B |
| Enrichment Score (ES) | -0.6125617 |
| Normalized Enrichment Score (NES) | -1.6476877 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.015237239 |
| FWER p-Value | 0.16 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ADAM19 | 5340075 | 59 | 2.212 | 0.0394 | No | ||
| 2 | ARID2 | 110647 5390435 | 108 | 1.650 | 0.0686 | No | ||
| 3 | HMGA1 | 6580408 | 507 | 0.743 | 0.0613 | No | ||
| 4 | VANGL2 | 70097 870075 | 567 | 0.683 | 0.0712 | No | ||
| 5 | FBXL19 | 4570039 | 778 | 0.554 | 0.0705 | No | ||
| 6 | ALS2CR2 | 6450128 7100092 | 954 | 0.479 | 0.0702 | No | ||
| 7 | ZDHHC18 | 6200059 | 1190 | 0.404 | 0.0652 | No | ||
| 8 | SH3KBP1 | 670102 2850458 7000309 | 1357 | 0.361 | 0.0631 | No | ||
| 9 | FGD1 | 4780021 | 1556 | 0.320 | 0.0586 | No | ||
| 10 | CELSR1 | 6510348 | 1594 | 0.313 | 0.0626 | No | ||
| 11 | EHD1 | 3840731 | 1685 | 0.295 | 0.0634 | No | ||
| 12 | ABHD2 | 4850504 6840010 | 1732 | 0.287 | 0.0664 | No | ||
| 13 | BAK1 | 6450563 | 2149 | 0.223 | 0.0481 | No | ||
| 14 | POM121 | 70152 | 2197 | 0.217 | 0.0497 | No | ||
| 15 | TMEM24 | 5860039 | 2200 | 0.217 | 0.0538 | No | ||
| 16 | BCR | 2260020 4230180 6040195 | 2229 | 0.213 | 0.0564 | No | ||
| 17 | CD200 | 940725 5260647 | 2290 | 0.202 | 0.0570 | No | ||
| 18 | ALDH5A1 | 7040750 | 2688 | 0.147 | 0.0382 | No | ||
| 19 | DEPDC1B | 2120458 | 2762 | 0.138 | 0.0369 | No | ||
| 20 | XKRX | 3800192 | 2777 | 0.136 | 0.0388 | No | ||
| 21 | CHFR | 3390050 6450300 6450736 | 2813 | 0.133 | 0.0395 | No | ||
| 22 | TRIB2 | 4120605 | 2828 | 0.131 | 0.0412 | No | ||
| 23 | SRP19 | 780450 3120075 | 3017 | 0.113 | 0.0332 | No | ||
| 24 | TMEM2 | 1980037 3290020 | 3035 | 0.111 | 0.0344 | No | ||
| 25 | MRAS | 2030524 | 3151 | 0.100 | 0.0301 | No | ||
| 26 | MXI1 | 5050064 5130484 | 3337 | 0.087 | 0.0217 | No | ||
| 27 | DLG5 | 450215 | 3381 | 0.084 | 0.0210 | No | ||
| 28 | KCNH7 | 7510068 | 3514 | 0.077 | 0.0153 | No | ||
| 29 | BLOC1S2 | 1240215 | 3572 | 0.074 | 0.0136 | No | ||
| 30 | SH3D19 | 6020520 | 3584 | 0.073 | 0.0144 | No | ||
| 31 | CSNK1G1 | 840082 1230575 3940647 | 3660 | 0.069 | 0.0117 | No | ||
| 32 | EIF3S10 | 1190079 3840176 | 3676 | 0.069 | 0.0122 | No | ||
| 33 | EPHA2 | 5890056 | 3888 | 0.060 | 0.0019 | No | ||
| 34 | CPSF2 | 6040709 | 4085 | 0.053 | -0.0077 | No | ||
| 35 | ALS2 | 3130546 6400070 | 4149 | 0.051 | -0.0102 | No | ||
| 36 | NARG1 | 5910563 6350095 | 4219 | 0.050 | -0.0130 | No | ||
| 37 | ST8SIA4 | 2680605 3060215 | 4240 | 0.049 | -0.0131 | No | ||
| 38 | PHF6 | 3120632 3990041 | 4335 | 0.046 | -0.0173 | No | ||
| 39 | FA2H | 730672 | 4513 | 0.042 | -0.0261 | No | ||
| 40 | TBC1D4 | 6420397 | 4649 | 0.038 | -0.0328 | No | ||
| 41 | PALMD | 2900369 | 4767 | 0.036 | -0.0384 | No | ||
| 42 | PTER | 3450685 | 4781 | 0.036 | -0.0384 | No | ||
| 43 | SALL1 | 5420020 7050195 | 4792 | 0.036 | -0.0383 | No | ||
| 44 | HPGD | 6770192 | 4817 | 0.035 | -0.0389 | No | ||
| 45 | PTPN13 | 2060681 | 4860 | 0.034 | -0.0405 | No | ||
| 46 | PLOD2 | 3360427 | 4967 | 0.033 | -0.0457 | No | ||
| 47 | COL10A1 | 2360307 | 5104 | 0.030 | -0.0525 | No | ||
| 48 | NOL4 | 5050446 5360519 | 5116 | 0.030 | -0.0525 | No | ||
| 49 | PIM1 | 630047 | 5187 | 0.028 | -0.0558 | No | ||
| 50 | WNK3 | 4060053 | 5360 | 0.026 | -0.0646 | No | ||
| 51 | RKHD3 | 6180471 | 5368 | 0.026 | -0.0645 | No | ||
| 52 | WNT5A | 840685 3120152 | 5387 | 0.025 | -0.0650 | No | ||
| 53 | AMOT | 1940121 2940647 6590093 | 5511 | 0.024 | -0.0712 | No | ||
| 54 | HTR2A | 360037 6330255 | 5512 | 0.024 | -0.0708 | No | ||
| 55 | VLDLR | 870722 3060047 5340452 6550131 | 5586 | 0.023 | -0.0743 | No | ||
| 56 | CACNA1C | 1660128 3780300 3870092 6290403 6900301 | 5718 | 0.022 | -0.0810 | No | ||
| 57 | PCK1 | 7000358 | 5722 | 0.022 | -0.0807 | No | ||
| 58 | STRBP | 4210594 5360239 | 5865 | 0.020 | -0.0880 | No | ||
| 59 | SENP5 | 4560537 6760465 | 5874 | 0.020 | -0.0881 | No | ||
| 60 | USP9X | 130435 2370735 3120338 | 5977 | 0.019 | -0.0933 | No | ||
| 61 | LIF | 1410373 2060129 3990338 | 6068 | 0.018 | -0.0978 | No | ||
| 62 | ZFHX4 | 6110167 | 6149 | 0.018 | -0.1018 | No | ||
| 63 | SYT10 | 5550048 | 6215 | 0.017 | -0.1050 | No | ||
| 64 | SLC4A4 | 1780075 1850040 4810068 | 6324 | 0.016 | -0.1106 | No | ||
| 65 | PHF21A | 610014 | 6503 | 0.015 | -0.1200 | No | ||
| 66 | HMGA2 | 2940121 3390647 5130279 6400136 | 6672 | 0.014 | -0.1289 | No | ||
| 67 | NAP1L5 | 360576 | 6698 | 0.013 | -0.1300 | No | ||
| 68 | LSM11 | 2350167 | 6700 | 0.013 | -0.1297 | No | ||
| 69 | ATP1A2 | 110278 | 6777 | 0.013 | -0.1336 | No | ||
| 70 | STYX | 1090358 | 6835 | 0.013 | -0.1365 | No | ||
| 71 | SPOCK2 | 6650369 | 6866 | 0.012 | -0.1379 | No | ||
| 72 | COL19A1 | 6220441 | 7113 | 0.011 | -0.1510 | No | ||
| 73 | SMAD1 | 630537 1850333 | 7237 | 0.011 | -0.1575 | No | ||
| 74 | MTX2 | 6660288 | 7544 | 0.009 | -0.1740 | No | ||
| 75 | SRGAP1 | 3940253 | 8004 | 0.008 | -0.1988 | No | ||
| 76 | ATM | 3610110 4050524 | 8120 | 0.007 | -0.2049 | No | ||
| 77 | ULK1 | 6100315 | 8230 | 0.007 | -0.2107 | No | ||
| 78 | KCNK1 | 1050068 | 8240 | 0.007 | -0.2111 | No | ||
| 79 | HOXA5 | 6840026 | 8322 | 0.006 | -0.2153 | No | ||
| 80 | TRPC3 | 840064 | 8390 | 0.006 | -0.2189 | No | ||
| 81 | USP25 | 3520035 | 8462 | 0.006 | -0.2226 | No | ||
| 82 | NUP50 | 1240519 2640390 2650471 4730133 | 8642 | 0.005 | -0.2322 | No | ||
| 83 | SLC24A4 | 2100487 | 8668 | 0.005 | -0.2335 | No | ||
| 84 | NTN4 | 6940398 | 8877 | 0.005 | -0.2447 | No | ||
| 85 | ATPAF1 | 2470164 | 9054 | 0.004 | -0.2542 | No | ||
| 86 | LRRC16 | 360022 6450070 | 9389 | 0.003 | -0.2723 | No | ||
| 87 | PURA | 3870156 | 9426 | 0.003 | -0.2742 | No | ||
| 88 | PNRC1 | 360025 | 9660 | 0.003 | -0.2868 | No | ||
| 89 | GABRB3 | 4150164 | 9729 | 0.002 | -0.2904 | No | ||
| 90 | JAG1 | 3440390 | 9772 | 0.002 | -0.2927 | No | ||
| 91 | PLEKHH1 | 4780280 | 9984 | 0.002 | -0.3041 | No | ||
| 92 | RCN1 | 2480041 | 10009 | 0.002 | -0.3054 | No | ||
| 93 | RPGR | 5910711 | 10213 | 0.001 | -0.3164 | No | ||
| 94 | HAS3 | 4200692 | 10231 | 0.001 | -0.3173 | No | ||
| 95 | MMP16 | 2680139 | 10269 | 0.001 | -0.3193 | No | ||
| 96 | NAB1 | 430739 1230114 3390736 6660056 | 10501 | 0.000 | -0.3318 | No | ||
| 97 | RCN2 | 840324 | 10556 | 0.000 | -0.3348 | No | ||
| 98 | KLF10 | 4850056 | 10681 | -0.000 | -0.3415 | No | ||
| 99 | SLC25A16 | 4200341 | 10780 | -0.000 | -0.3468 | No | ||
| 100 | ZDHHC6 | 510600 | 10805 | -0.000 | -0.3481 | No | ||
| 101 | NFE2L3 | 780100 | 10965 | -0.001 | -0.3568 | No | ||
| 102 | POF1B | 3800670 | 10985 | -0.001 | -0.3578 | No | ||
| 103 | BTBD7 | 730288 | 10997 | -0.001 | -0.3584 | No | ||
| 104 | ZNF462 | 6900100 | 11175 | -0.001 | -0.3680 | No | ||
| 105 | ACVR1C | 5890044 | 11365 | -0.002 | -0.3782 | No | ||
| 106 | USP15 | 610592 3520504 | 11378 | -0.002 | -0.3788 | No | ||
| 107 | DCDC2 | 7000338 | 11387 | -0.002 | -0.3792 | No | ||
| 108 | KCNQ4 | 3800438 7050400 | 11541 | -0.002 | -0.3875 | No | ||
| 109 | CLASP2 | 2510139 | 11663 | -0.003 | -0.3940 | No | ||
| 110 | HGF | 3360593 | 11672 | -0.003 | -0.3944 | No | ||
| 111 | PDHX | 870315 | 11697 | -0.003 | -0.3957 | No | ||
| 112 | PHLDB2 | 6590685 | 11833 | -0.003 | -0.4029 | No | ||
| 113 | PCDH18 | 430010 | 12045 | -0.004 | -0.4143 | No | ||
| 114 | MITF | 380056 | 12085 | -0.004 | -0.4164 | No | ||
| 115 | HAO1 | 7050093 | 12227 | -0.004 | -0.4240 | No | ||
| 116 | FANCA | 6130070 | 12339 | -0.005 | -0.4299 | No | ||
| 117 | UBE2D1 | 1850400 | 12396 | -0.005 | -0.4328 | No | ||
| 118 | RAP1A | 1090025 | 12445 | -0.005 | -0.4354 | No | ||
| 119 | ASPN | 2450021 | 12562 | -0.006 | -0.4416 | No | ||
| 120 | RGS4 | 2970711 | 12646 | -0.006 | -0.4459 | No | ||
| 121 | MAB21L1 | 1450634 1580092 | 12790 | -0.007 | -0.4536 | No | ||
| 122 | LGR4 | 1660309 | 12841 | -0.007 | -0.4562 | No | ||
| 123 | PLCB1 | 2570050 7100102 | 12861 | -0.007 | -0.4571 | No | ||
| 124 | PTGS2 | 2510301 3170369 | 12973 | -0.008 | -0.4630 | No | ||
| 125 | PELI2 | 1940364 | 13216 | -0.009 | -0.4759 | No | ||
| 126 | ACSL3 | 3140195 | 13498 | -0.011 | -0.4910 | No | ||
| 127 | SSX2IP | 4610176 | 13554 | -0.011 | -0.4938 | No | ||
| 128 | PAPD4 | 1580673 730605 5270112 | 13559 | -0.011 | -0.4938 | No | ||
| 129 | KCNJ2 | 630019 | 13584 | -0.011 | -0.4949 | No | ||
| 130 | PBEF1 | 1090278 | 13761 | -0.013 | -0.5042 | No | ||
| 131 | NDFIP2 | 4070451 | 13771 | -0.013 | -0.5044 | No | ||
| 132 | ACBD5 | 3440397 6020600 | 13836 | -0.014 | -0.5076 | No | ||
| 133 | DAPK1 | 5570010 | 13837 | -0.014 | -0.5074 | No | ||
| 134 | ITGA5 | 5550520 | 13894 | -0.015 | -0.5101 | No | ||
| 135 | MARK1 | 450484 | 14010 | -0.016 | -0.5161 | No | ||
| 136 | ATP11C | 770672 7040129 | 14014 | -0.016 | -0.5159 | No | ||
| 137 | STAC2 | 7100022 | 14024 | -0.016 | -0.5161 | No | ||
| 138 | E2F7 | 4200056 | 14044 | -0.017 | -0.5168 | No | ||
| 139 | RTN1 | 4150452 4850176 | 14138 | -0.018 | -0.5215 | No | ||
| 140 | CAMSAP1 | 1580131 2120494 6620446 | 14179 | -0.019 | -0.5233 | No | ||
| 141 | PCDH9 | 2100136 | 14192 | -0.019 | -0.5236 | No | ||
| 142 | PTP4A1 | 130692 | 14223 | -0.019 | -0.5249 | No | ||
| 143 | SSH2 | 360056 | 14252 | -0.020 | -0.5260 | No | ||
| 144 | APC | 3850484 5860722 | 14661 | -0.030 | -0.5476 | No | ||
| 145 | STK39 | 50692 | 14681 | -0.031 | -0.5481 | No | ||
| 146 | SLC1A1 | 520372 6420059 | 14694 | -0.031 | -0.5481 | No | ||
| 147 | NEK6 | 3360687 | 14712 | -0.032 | -0.5484 | No | ||
| 148 | PITPNC1 | 3990017 | 14771 | -0.034 | -0.5509 | No | ||
| 149 | CDK6 | 4920253 | 14772 | -0.034 | -0.5503 | No | ||
| 150 | LTBP1 | 780035 2100746 2760402 3780551 6550184 6860193 | 14802 | -0.035 | -0.5512 | No | ||
| 151 | PDGFRA | 2940332 | 14824 | -0.036 | -0.5516 | No | ||
| 152 | VAMP1 | 450735 1940707 | 15135 | -0.051 | -0.5675 | No | ||
| 153 | BRWD1 | 3130048 3140168 6400601 | 15171 | -0.052 | -0.5684 | No | ||
| 154 | NRIP1 | 3190039 | 15228 | -0.055 | -0.5704 | No | ||
| 155 | PAWR | 5130546 6330706 | 15237 | -0.056 | -0.5697 | No | ||
| 156 | ATF2 | 2260348 2480441 | 15266 | -0.057 | -0.5702 | No | ||
| 157 | SLC12A2 | 4610019 6040672 | 15470 | -0.070 | -0.5798 | No | ||
| 158 | IGF1 | 1990193 3130377 3290280 | 15490 | -0.071 | -0.5795 | No | ||
| 159 | RPS6KA2 | 2970161 4730278 | 15497 | -0.072 | -0.5784 | No | ||
| 160 | AMPH | 2260338 3140576 | 15501 | -0.072 | -0.5772 | No | ||
| 161 | YTHDF3 | 4540632 4670717 7040609 | 15571 | -0.077 | -0.5795 | No | ||
| 162 | THRAP2 | 4920600 | 15592 | -0.079 | -0.5791 | No | ||
| 163 | USP37 | 2470433 | 15707 | -0.087 | -0.5836 | No | ||
| 164 | ACADSB | 520170 | 15949 | -0.110 | -0.5945 | No | ||
| 165 | COX5A | 1450609 | 16009 | -0.118 | -0.5955 | No | ||
| 166 | CDK2AP1 | 2340156 | 16020 | -0.119 | -0.5937 | No | ||
| 167 | ULK2 | 50041 5050176 | 16228 | -0.144 | -0.6022 | No | ||
| 168 | TAF12 | 3710746 4920300 6180603 | 16258 | -0.147 | -0.6009 | No | ||
| 169 | ARL4C | 1190279 | 16299 | -0.154 | -0.6001 | No | ||
| 170 | PFKFB3 | 630706 | 16400 | -0.168 | -0.6023 | No | ||
| 171 | CCNE1 | 110064 | 16589 | -0.205 | -0.6086 | Yes | ||
| 172 | YPEL1 | 6590435 | 16599 | -0.206 | -0.6051 | Yes | ||
| 173 | MIB1 | 3800537 | 16713 | -0.232 | -0.6068 | Yes | ||
| 174 | USP3 | 2060332 | 16754 | -0.242 | -0.6043 | Yes | ||
| 175 | RTF1 | 2100735 | 16811 | -0.256 | -0.6024 | Yes | ||
| 176 | AMMECR1 | 3800435 | 16894 | -0.274 | -0.6016 | Yes | ||
| 177 | BHLHB2 | 7040603 | 16960 | -0.290 | -0.5995 | Yes | ||
| 178 | MKNK2 | 1240020 5270092 | 17140 | -0.341 | -0.6027 | Yes | ||
| 179 | OSBPL2 | 5690215 | 17169 | -0.349 | -0.5975 | Yes | ||
| 180 | SLC38A2 | 1470242 3800026 | 17287 | -0.378 | -0.5966 | Yes | ||
| 181 | ZFX | 5900400 | 17341 | -0.399 | -0.5918 | Yes | ||
| 182 | RANBP10 | 2340184 | 17379 | -0.413 | -0.5858 | Yes | ||
| 183 | PAK2 | 360438 7050068 | 17382 | -0.414 | -0.5780 | Yes | ||
| 184 | EPC2 | 2470095 | 17424 | -0.427 | -0.5720 | Yes | ||
| 185 | GSK3B | 5360348 | 17444 | -0.435 | -0.5646 | Yes | ||
| 186 | SLC4A7 | 2370056 | 17468 | -0.445 | -0.5573 | Yes | ||
| 187 | PRKCD | 770592 | 17479 | -0.447 | -0.5492 | Yes | ||
| 188 | PHTF2 | 6590053 | 17509 | -0.458 | -0.5420 | Yes | ||
| 189 | SLC7A11 | 2850138 | 17542 | -0.470 | -0.5347 | Yes | ||
| 190 | CUGBP2 | 6180121 6840139 | 17550 | -0.471 | -0.5260 | Yes | ||
| 191 | OSBPL11 | 1770044 | 17574 | -0.486 | -0.5178 | Yes | ||
| 192 | EZH2 | 6130605 6380524 | 17642 | -0.510 | -0.5117 | Yes | ||
| 193 | TMEM33 | 2340022 3940750 | 17677 | -0.525 | -0.5034 | Yes | ||
| 194 | MAN2A1 | 6650176 | 17688 | -0.530 | -0.4937 | Yes | ||
| 195 | GTF3C2 | 670494 | 17689 | -0.531 | -0.4835 | Yes | ||
| 196 | ARPP-19 | 5700025 | 17775 | -0.564 | -0.4772 | Yes | ||
| 197 | CMTM4 | 360600 3520332 | 17834 | -0.595 | -0.4689 | Yes | ||
| 198 | SRPK1 | 450110 | 17839 | -0.596 | -0.4577 | Yes | ||
| 199 | ADM | 4810121 | 17841 | -0.597 | -0.4462 | Yes | ||
| 200 | TOB1 | 4150138 | 17941 | -0.646 | -0.4391 | Yes | ||
| 201 | KPNA2 | 3450114 | 17944 | -0.648 | -0.4268 | Yes | ||
| 202 | UBE2G1 | 2230546 5420528 7040463 | 18223 | -0.851 | -0.4255 | Yes | ||
| 203 | SMAD4 | 5670519 | 18224 | -0.852 | -0.4091 | Yes | ||
| 204 | MAPK6 | 6760520 | 18240 | -0.875 | -0.3930 | Yes | ||
| 205 | TTC13 | 3520750 | 18249 | -0.886 | -0.3764 | Yes | ||
| 206 | RAP2C | 1690132 | 18260 | -0.898 | -0.3596 | Yes | ||
| 207 | ENC1 | 4810348 | 18327 | -0.976 | -0.3444 | Yes | ||
| 208 | EP400 | 3940722 | 18331 | -0.983 | -0.3256 | Yes | ||
| 209 | MOBK1B | 4810373 | 18338 | -0.987 | -0.3070 | Yes | ||
| 210 | INHBB | 2680092 | 18351 | -1.007 | -0.2882 | Yes | ||
| 211 | RNF6 | 2640035 | 18353 | -1.011 | -0.2688 | Yes | ||
| 212 | MCL1 | 1660672 | 18416 | -1.131 | -0.2504 | Yes | ||
| 213 | PPP3R1 | 1190041 1570487 | 18466 | -1.284 | -0.2283 | Yes | ||
| 214 | EIF5 | 6380750 | 18467 | -1.296 | -0.2033 | Yes | ||
| 215 | ARHGAP17 | 4810095 5360563 5900427 | 18490 | -1.372 | -0.1781 | Yes | ||
| 216 | TNRC6A | 1240537 6130037 6200010 | 18507 | -1.429 | -0.1514 | Yes | ||
| 217 | EIF4G2 | 3800575 6860184 | 18522 | -1.497 | -0.1234 | Yes | ||
| 218 | CBFB | 1850551 2650035 | 18525 | -1.525 | -0.0941 | Yes | ||
| 219 | PPP3CB | 6020156 | 18526 | -1.527 | -0.0647 | Yes | ||
| 220 | CCND2 | 5340167 | 18543 | -1.609 | -0.0345 | Yes | ||
| 221 | UBE4B | 780008 3610154 | 18570 | -1.996 | 0.0025 | Yes |

