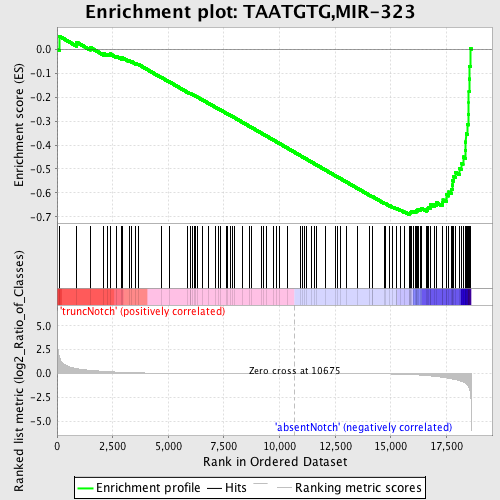
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #truncNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#truncNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | TAATGTG,MIR-323 |
| Enrichment Score (ES) | -0.6897647 |
| Normalized Enrichment Score (NES) | -1.7071809 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.007839775 |
| FWER p-Value | 0.045 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SPSB4 | 4070673 | 95 | 1.753 | 0.0541 | No | ||
| 2 | PTK2B | 4730411 | 875 | 0.505 | 0.0291 | No | ||
| 3 | CCND1 | 460524 770309 3120576 6980398 | 1487 | 0.331 | 0.0072 | No | ||
| 4 | MAP3K3 | 610685 | 2091 | 0.231 | -0.0175 | No | ||
| 5 | FBN1 | 3170181 | 2284 | 0.204 | -0.0210 | No | ||
| 6 | ACAT2 | 2680400 3610440 | 2379 | 0.189 | -0.0197 | No | ||
| 7 | NFKB1 | 5420358 | 2660 | 0.151 | -0.0297 | No | ||
| 8 | TOP1 | 770471 4060632 6650324 | 2912 | 0.123 | -0.0391 | No | ||
| 9 | TMEM16D | 2650324 | 2926 | 0.121 | -0.0358 | No | ||
| 10 | ZNF318 | 5890091 6130021 | 3235 | 0.094 | -0.0492 | No | ||
| 11 | EGR3 | 6940128 | 3356 | 0.086 | -0.0528 | No | ||
| 12 | CDKN1B | 3800025 6450044 | 3544 | 0.076 | -0.0603 | No | ||
| 13 | PPP1CB | 3130411 3780270 4150180 | 3643 | 0.070 | -0.0633 | No | ||
| 14 | UHRF2 | 60050 840280 1340746 5080091 | 4680 | 0.038 | -0.1180 | No | ||
| 15 | STAT5B | 6200026 | 5048 | 0.031 | -0.1367 | No | ||
| 16 | OSBP | 5390471 | 5864 | 0.020 | -0.1801 | No | ||
| 17 | USP9X | 130435 2370735 3120338 | 5977 | 0.019 | -0.1855 | No | ||
| 18 | CXCL12 | 580546 4150750 4570068 | 5997 | 0.019 | -0.1859 | No | ||
| 19 | POU2F3 | 2030601 | 6073 | 0.018 | -0.1893 | No | ||
| 20 | PKP4 | 3710400 5890097 | 6164 | 0.017 | -0.1936 | No | ||
| 21 | HOXB5 | 2900538 | 6224 | 0.017 | -0.1962 | No | ||
| 22 | SLC4A4 | 1780075 1850040 4810068 | 6324 | 0.016 | -0.2010 | No | ||
| 23 | RAB11FIP2 | 4570040 | 6518 | 0.015 | -0.2110 | No | ||
| 24 | TMOD1 | 3850100 | 6784 | 0.013 | -0.2248 | No | ||
| 25 | STK35 | 2360121 | 7110 | 0.011 | -0.2420 | No | ||
| 26 | GPM6A | 1660044 2750152 | 7246 | 0.011 | -0.2490 | No | ||
| 27 | ZIC1 | 670113 | 7354 | 0.010 | -0.2544 | No | ||
| 28 | SEPT3 | 3830022 5290079 | 7624 | 0.009 | -0.2686 | No | ||
| 29 | PAK7 | 3830164 | 7655 | 0.009 | -0.2700 | No | ||
| 30 | EIF3S1 | 6130368 6770044 | 7811 | 0.008 | -0.2781 | No | ||
| 31 | FBN2 | 50435 | 7862 | 0.008 | -0.2805 | No | ||
| 32 | SEMA6D | 4050324 5860138 6350307 | 7954 | 0.008 | -0.2851 | No | ||
| 33 | KCNJ3 | 50184 | 8350 | 0.006 | -0.3063 | No | ||
| 34 | DPF2 | 6980373 | 8634 | 0.005 | -0.3214 | No | ||
| 35 | LPP | 1660541 | 8740 | 0.005 | -0.3269 | No | ||
| 36 | STX12 | 610451 | 9193 | 0.004 | -0.3512 | No | ||
| 37 | CLCN3 | 510195 780176 1660093 3830372 | 9290 | 0.003 | -0.3562 | No | ||
| 38 | MYLIP | 50717 4050672 | 9423 | 0.003 | -0.3633 | No | ||
| 39 | NRK | 4590609 | 9726 | 0.002 | -0.3795 | No | ||
| 40 | MCFD2 | 520576 5690017 | 9873 | 0.002 | -0.3873 | No | ||
| 41 | RCN1 | 2480041 | 10009 | 0.002 | -0.3946 | No | ||
| 42 | BET1 | 4010725 | 10338 | 0.001 | -0.4123 | No | ||
| 43 | CRK | 1230162 4780128 | 10920 | -0.001 | -0.4436 | No | ||
| 44 | EBF3 | 4200129 | 11023 | -0.001 | -0.4491 | No | ||
| 45 | QSCN6L1 | 870040 | 11126 | -0.001 | -0.4546 | No | ||
| 46 | TGFA | 1570332 | 11226 | -0.001 | -0.4599 | No | ||
| 47 | AUH | 5570152 | 11415 | -0.002 | -0.4700 | No | ||
| 48 | TEAD1 | 2470551 | 11588 | -0.002 | -0.4792 | No | ||
| 49 | HOXA1 | 1190524 5420142 | 11643 | -0.003 | -0.4820 | No | ||
| 50 | WTAP | 4010176 | 12056 | -0.004 | -0.5042 | No | ||
| 51 | LRRC4 | 3850041 | 12521 | -0.006 | -0.5290 | No | ||
| 52 | FGD4 | 520168 870411 2640253 6550338 6650364 | 12585 | -0.006 | -0.5322 | No | ||
| 53 | POU4F2 | 2120195 2570022 | 12715 | -0.006 | -0.5390 | No | ||
| 54 | RSBN1 | 7000487 | 12985 | -0.008 | -0.5533 | No | ||
| 55 | TBPL1 | 3610332 | 13522 | -0.011 | -0.5819 | No | ||
| 56 | NR4A3 | 2900021 5860095 5910039 | 14052 | -0.017 | -0.6099 | No | ||
| 57 | PPARGC1A | 4670040 | 14182 | -0.019 | -0.6162 | No | ||
| 58 | PCDH9 | 2100136 | 14192 | -0.019 | -0.6161 | No | ||
| 59 | ANKRD27 | 2190465 4070300 6040026 | 14716 | -0.032 | -0.6433 | No | ||
| 60 | SLITRK2 | 2640020 | 14757 | -0.033 | -0.6443 | No | ||
| 61 | HOOK3 | 1190242 2470059 | 14961 | -0.042 | -0.6539 | No | ||
| 62 | CREB1 | 1500717 2230358 3610600 6550601 | 15093 | -0.048 | -0.6593 | No | ||
| 63 | TOP2B | 6980309 | 15235 | -0.056 | -0.6650 | No | ||
| 64 | DCUN1D1 | 2360332 | 15243 | -0.056 | -0.6635 | No | ||
| 65 | ATP11B | 4230100 | 15438 | -0.068 | -0.6717 | No | ||
| 66 | NFAT5 | 2510411 5890195 6550152 | 15629 | -0.081 | -0.6792 | No | ||
| 67 | ASF1A | 7050020 | 15825 | -0.098 | -0.6865 | Yes | ||
| 68 | TRIM33 | 580619 2230280 3990433 6200747 | 15841 | -0.099 | -0.6839 | Yes | ||
| 69 | ZHX1 | 2100110 2120286 | 15866 | -0.101 | -0.6818 | Yes | ||
| 70 | RAB10 | 2360008 5890020 | 15894 | -0.105 | -0.6797 | Yes | ||
| 71 | PDCD10 | 4760500 | 15915 | -0.107 | -0.6772 | Yes | ||
| 72 | HIPK1 | 110193 | 16015 | -0.118 | -0.6785 | Yes | ||
| 73 | KPNA3 | 7040088 | 16100 | -0.126 | -0.6788 | Yes | ||
| 74 | CLASP1 | 6860279 | 16134 | -0.130 | -0.6762 | Yes | ||
| 75 | RPS6KA3 | 1980707 | 16162 | -0.134 | -0.6731 | Yes | ||
| 76 | MAP4K4 | 2360059 | 16207 | -0.141 | -0.6708 | Yes | ||
| 77 | ULK2 | 50041 5050176 | 16228 | -0.144 | -0.6670 | Yes | ||
| 78 | REV3L | 1090717 | 16350 | -0.161 | -0.6681 | Yes | ||
| 79 | MRC1 | 730097 | 16378 | -0.164 | -0.6640 | Yes | ||
| 80 | PJA2 | 430537 | 16596 | -0.206 | -0.6687 | Yes | ||
| 81 | WBP2 | 3170128 | 16665 | -0.221 | -0.6649 | Yes | ||
| 82 | HS2ST1 | 3170088 | 16692 | -0.229 | -0.6586 | Yes | ||
| 83 | ZDHHC21 | 2370338 | 16788 | -0.251 | -0.6553 | Yes | ||
| 84 | CSNK1G3 | 110450 | 16790 | -0.251 | -0.6468 | Yes | ||
| 85 | CEPT1 | 1940047 | 16977 | -0.295 | -0.6469 | Yes | ||
| 86 | SMARCAD1 | 5390619 | 17062 | -0.322 | -0.6406 | Yes | ||
| 87 | CUL1 | 1990632 | 17308 | -0.386 | -0.6408 | Yes | ||
| 88 | FEM1B | 1230110 | 17344 | -0.400 | -0.6292 | Yes | ||
| 89 | CHCHD4 | 2680075 | 17493 | -0.453 | -0.6218 | Yes | ||
| 90 | KPNA1 | 5270324 | 17494 | -0.453 | -0.6065 | Yes | ||
| 91 | UTX | 2900017 6770452 | 17610 | -0.499 | -0.5959 | Yes | ||
| 92 | DHX36 | 2470465 | 17729 | -0.548 | -0.5838 | Yes | ||
| 93 | ARFIP1 | 3130601 | 17762 | -0.560 | -0.5665 | Yes | ||
| 94 | SUMO1 | 1990364 2760338 6380195 | 17780 | -0.568 | -0.5483 | Yes | ||
| 95 | RAB1A | 2370671 | 17825 | -0.589 | -0.5308 | Yes | ||
| 96 | RANBP9 | 4670685 | 17903 | -0.626 | -0.5138 | Yes | ||
| 97 | DDX3X | 2190020 | 18098 | -0.751 | -0.4988 | Yes | ||
| 98 | TMPO | 4050494 | 18166 | -0.804 | -0.4753 | Yes | ||
| 99 | NDEL1 | 2370465 | 18246 | -0.881 | -0.4498 | Yes | ||
| 100 | MAPRE1 | 3290037 | 18349 | -1.005 | -0.4214 | Yes | ||
| 101 | NFIX | 2450152 | 18366 | -1.032 | -0.3874 | Yes | ||
| 102 | PITPNA | 2760332 | 18379 | -1.048 | -0.3526 | Yes | ||
| 103 | UBL3 | 6450458 | 18448 | -1.221 | -0.3150 | Yes | ||
| 104 | SATB1 | 5670154 | 18479 | -1.341 | -0.2713 | Yes | ||
| 105 | TNRC6A | 1240537 6130037 6200010 | 18507 | -1.429 | -0.2245 | Yes | ||
| 106 | SFRS1 | 2360440 | 18511 | -1.447 | -0.1757 | Yes | ||
| 107 | ENTPD5 | 4280070 | 18528 | -1.538 | -0.1246 | Yes | ||
| 108 | SLMAP | 3120093 4760128 5390528 | 18542 | -1.608 | -0.0710 | Yes | ||
| 109 | GJA1 | 5220731 | 18585 | -2.218 | 0.0017 | Yes |

