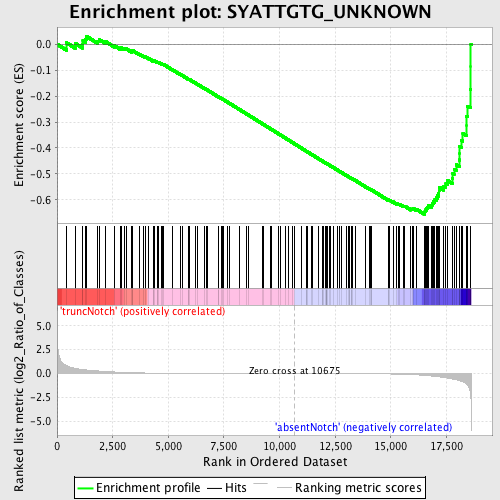
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #truncNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#truncNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | SYATTGTG_UNKNOWN |
| Enrichment Score (ES) | -0.6564448 |
| Normalized Enrichment Score (NES) | -1.6864637 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.009989331 |
| FWER p-Value | 0.075 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SIRT6 | 540731 1400735 | 421 | 0.832 | 0.0060 | No | ||
| 2 | RABAC1 | 2030435 2100632 | 813 | 0.535 | 0.0034 | No | ||
| 3 | CHD4 | 5420059 6130338 6380717 | 1150 | 0.415 | -0.0005 | No | ||
| 4 | OTUB1 | 610551 | 1160 | 0.412 | 0.0133 | No | ||
| 5 | HSPB2 | 1990110 | 1281 | 0.378 | 0.0199 | No | ||
| 6 | TLE3 | 580040 4730121 | 1308 | 0.372 | 0.0314 | No | ||
| 7 | ITPR3 | 4010632 | 1836 | 0.270 | 0.0122 | No | ||
| 8 | SF4 | 3830341 | 1885 | 0.261 | 0.0186 | No | ||
| 9 | TOMM40 | 6110017 | 2184 | 0.219 | 0.0101 | No | ||
| 10 | NFIL3 | 4070377 | 2581 | 0.163 | -0.0057 | No | ||
| 11 | POFUT1 | 1570458 | 2843 | 0.129 | -0.0154 | No | ||
| 12 | IFRD2 | 4150110 | 2888 | 0.126 | -0.0134 | No | ||
| 13 | PDYN | 5720408 | 3029 | 0.112 | -0.0171 | No | ||
| 14 | TPM3 | 670600 3170296 5670167 6650471 | 3101 | 0.104 | -0.0174 | No | ||
| 15 | EGR3 | 6940128 | 3356 | 0.086 | -0.0281 | No | ||
| 16 | GRIA3 | 2900164 6130278 6860154 | 3378 | 0.085 | -0.0263 | No | ||
| 17 | ETV4 | 2450605 | 3387 | 0.084 | -0.0238 | No | ||
| 18 | EXT1 | 4540603 | 3707 | 0.067 | -0.0388 | No | ||
| 19 | PYGO1 | 840575 | 3870 | 0.061 | -0.0455 | No | ||
| 20 | CBLB | 3520465 4670348 | 3955 | 0.058 | -0.0480 | No | ||
| 21 | BCL11A | 6860369 | 4121 | 0.052 | -0.0551 | No | ||
| 22 | PRDM13 | 1430022 | 4338 | 0.046 | -0.0652 | No | ||
| 23 | FOXP2 | 3520561 4150372 4760524 | 4343 | 0.046 | -0.0638 | No | ||
| 24 | REC8L1 | 5900014 | 4370 | 0.045 | -0.0637 | No | ||
| 25 | PRIMA1 | 1500673 6760671 | 4505 | 0.042 | -0.0695 | No | ||
| 26 | ACRV1 | 4070161 | 4533 | 0.041 | -0.0695 | No | ||
| 27 | RNF38 | 1940746 3140133 3840601 | 4560 | 0.040 | -0.0695 | No | ||
| 28 | SLC36A3 | 430075 | 4691 | 0.037 | -0.0752 | No | ||
| 29 | HOXC6 | 3850133 4920736 | 4742 | 0.036 | -0.0767 | No | ||
| 30 | PPP2R5C | 3310121 6650672 | 4802 | 0.035 | -0.0787 | No | ||
| 31 | GRIK3 | 6380592 | 5165 | 0.029 | -0.0973 | No | ||
| 32 | SCUBE2 | 2850315 5130040 | 5548 | 0.024 | -0.1171 | No | ||
| 33 | CLDN17 | 3710672 | 5628 | 0.023 | -0.1206 | No | ||
| 34 | HOXC4 | 1940193 | 5896 | 0.020 | -0.1344 | No | ||
| 35 | CELSR3 | 5270731 7100082 | 5914 | 0.020 | -0.1346 | No | ||
| 36 | CSNK2B | 5080161 | 5950 | 0.019 | -0.1358 | No | ||
| 37 | CA3 | 870687 5890390 | 6198 | 0.017 | -0.1486 | No | ||
| 38 | MB | 730019 | 6302 | 0.016 | -0.1536 | No | ||
| 39 | IBSP | 2060048 | 6606 | 0.014 | -0.1695 | No | ||
| 40 | NFIB | 460450 | 6719 | 0.013 | -0.1751 | No | ||
| 41 | GJA12 | 430273 4560731 | 6769 | 0.013 | -0.1773 | No | ||
| 42 | SMAD1 | 630537 1850333 | 7237 | 0.011 | -0.2022 | No | ||
| 43 | GPM6A | 1660044 2750152 | 7246 | 0.011 | -0.2023 | No | ||
| 44 | RFX4 | 3710100 5340215 | 7401 | 0.010 | -0.2103 | No | ||
| 45 | MYH2 | 2340433 | 7455 | 0.009 | -0.2128 | No | ||
| 46 | PITX2 | 870537 2690139 | 7494 | 0.009 | -0.2146 | No | ||
| 47 | TRIB1 | 2320435 | 7666 | 0.009 | -0.2235 | No | ||
| 48 | DPYSL3 | 1580594 | 7759 | 0.008 | -0.2282 | No | ||
| 49 | RAB3C | 5420072 | 8193 | 0.007 | -0.2514 | No | ||
| 50 | CBX8 | 50671 | 8491 | 0.006 | -0.2673 | No | ||
| 51 | FMR1 | 5050075 | 8609 | 0.006 | -0.2734 | No | ||
| 52 | FGF14 | 2630390 3520075 6770048 | 9231 | 0.004 | -0.3069 | No | ||
| 53 | LAMA1 | 1770594 | 9283 | 0.004 | -0.3096 | No | ||
| 54 | KIF21A | 2970338 | 9591 | 0.003 | -0.3261 | No | ||
| 55 | PAX3 | 50551 | 9649 | 0.003 | -0.3291 | No | ||
| 56 | PCSK1N | 4010435 | 9933 | 0.002 | -0.3443 | No | ||
| 57 | PCDH8 | 2360047 4200452 | 10061 | 0.002 | -0.3512 | No | ||
| 58 | MMP16 | 2680139 | 10269 | 0.001 | -0.3623 | No | ||
| 59 | REST | 1500471 | 10387 | 0.001 | -0.3686 | No | ||
| 60 | FEZ1 | 2630725 4810324 | 10557 | 0.000 | -0.3778 | No | ||
| 61 | FGF17 | 3130022 | 10652 | 0.000 | -0.3829 | No | ||
| 62 | PVRL3 | 1400324 4200593 6650128 | 11003 | -0.001 | -0.4018 | No | ||
| 63 | ADAM22 | 5390368 | 11194 | -0.001 | -0.4120 | No | ||
| 64 | CFL2 | 380239 6200368 | 11200 | -0.001 | -0.4122 | No | ||
| 65 | MEIS1 | 1400575 | 11234 | -0.001 | -0.4140 | No | ||
| 66 | NNAT | 4920053 5130253 | 11413 | -0.002 | -0.4235 | No | ||
| 67 | FSCN3 | 60093 | 11465 | -0.002 | -0.4262 | No | ||
| 68 | BAT4 | 2690128 6220451 | 11765 | -0.003 | -0.4423 | No | ||
| 69 | UCN2 | 4590372 | 11910 | -0.003 | -0.4500 | No | ||
| 70 | OMG | 6760066 | 11937 | -0.003 | -0.4513 | No | ||
| 71 | PLXNC1 | 5860315 | 11974 | -0.004 | -0.4531 | No | ||
| 72 | TSNAXIP1 | 1770270 6180577 | 12050 | -0.004 | -0.4570 | No | ||
| 73 | SLITRK4 | 5910022 | 12100 | -0.004 | -0.4595 | No | ||
| 74 | LIG1 | 1980438 4540112 5570609 | 12106 | -0.004 | -0.4597 | No | ||
| 75 | ARHGEF19 | 940102 | 12123 | -0.004 | -0.4604 | No | ||
| 76 | GSH1 | 2760242 | 12141 | -0.004 | -0.4612 | No | ||
| 77 | CCNT2 | 2100390 4150563 | 12259 | -0.005 | -0.4673 | No | ||
| 78 | GRIN2B | 3800333 | 12306 | -0.005 | -0.4697 | No | ||
| 79 | DLL1 | 1770377 | 12429 | -0.005 | -0.4761 | No | ||
| 80 | BSN | 5270347 | 12592 | -0.006 | -0.4847 | No | ||
| 81 | TNP1 | 3120048 | 12707 | -0.006 | -0.4906 | No | ||
| 82 | MYL3 | 6040563 | 12787 | -0.007 | -0.4947 | No | ||
| 83 | NNT | 540253 1170471 5550092 6760397 | 12998 | -0.008 | -0.5058 | No | ||
| 84 | LIN28 | 6590672 | 13088 | -0.008 | -0.5103 | No | ||
| 85 | ASB17 | 3520433 | 13147 | -0.008 | -0.5131 | No | ||
| 86 | OPHN1 | 2360100 | 13162 | -0.008 | -0.5136 | No | ||
| 87 | CLDN5 | 6510717 | 13246 | -0.009 | -0.5178 | No | ||
| 88 | SEMA3A | 1780064 | 13249 | -0.009 | -0.5176 | No | ||
| 89 | SH3BGRL2 | 1780239 | 13289 | -0.009 | -0.5194 | No | ||
| 90 | HOXA2 | 2120121 | 13430 | -0.010 | -0.5266 | No | ||
| 91 | ERG | 50154 1770739 | 13880 | -0.014 | -0.5504 | No | ||
| 92 | NR4A3 | 2900021 5860095 5910039 | 14052 | -0.017 | -0.5591 | No | ||
| 93 | HOXA7 | 5910152 | 14071 | -0.017 | -0.5595 | No | ||
| 94 | RTN1 | 4150452 4850176 | 14138 | -0.018 | -0.5624 | No | ||
| 95 | GALC | 2260685 4810161 6370040 | 14151 | -0.018 | -0.5624 | No | ||
| 96 | CA2 | 1660113 1660600 | 14877 | -0.038 | -0.6004 | No | ||
| 97 | THAP11 | 6940025 | 14883 | -0.038 | -0.5993 | No | ||
| 98 | CASK | 2340215 3290576 6770215 | 14939 | -0.041 | -0.6009 | No | ||
| 99 | OLFML3 | 7100463 | 15100 | -0.049 | -0.6079 | No | ||
| 100 | BAG5 | 2230368 6420537 | 15269 | -0.058 | -0.6150 | No | ||
| 101 | ATE1 | 290491 6940722 | 15330 | -0.061 | -0.6161 | No | ||
| 102 | CAMK2A | 1740333 1940112 4150292 | 15377 | -0.064 | -0.6164 | No | ||
| 103 | YTHDF3 | 4540632 4670717 7040609 | 15571 | -0.077 | -0.6242 | No | ||
| 104 | GUCA1A | 6020338 | 15618 | -0.080 | -0.6239 | No | ||
| 105 | NDST2 | 6290711 | 15905 | -0.106 | -0.6357 | No | ||
| 106 | RNGTT | 780152 780746 1090471 | 15967 | -0.113 | -0.6351 | No | ||
| 107 | MYO1C | 3190398 | 16013 | -0.118 | -0.6334 | No | ||
| 108 | POLR3E | 380152 | 16146 | -0.131 | -0.6360 | No | ||
| 109 | NSD1 | 4560519 4780672 | 16524 | -0.193 | -0.6498 | Yes | ||
| 110 | YWHAZ | 1230717 | 16534 | -0.195 | -0.6435 | Yes | ||
| 111 | ORMDL3 | 1340711 | 16576 | -0.201 | -0.6388 | Yes | ||
| 112 | PLAGL2 | 1770148 | 16594 | -0.206 | -0.6326 | Yes | ||
| 113 | CLU | 5420075 | 16642 | -0.216 | -0.6276 | Yes | ||
| 114 | RPL30 | 4810369 | 16698 | -0.231 | -0.6226 | Yes | ||
| 115 | ARIH1 | 6900025 | 16829 | -0.260 | -0.6207 | Yes | ||
| 116 | MAPK10 | 6110193 | 16856 | -0.267 | -0.6128 | Yes | ||
| 117 | DLX1 | 360168 | 16912 | -0.279 | -0.6061 | Yes | ||
| 118 | BCL2L1 | 1580452 4200152 5420484 | 16980 | -0.296 | -0.5995 | Yes | ||
| 119 | BCL7A | 4560095 | 17047 | -0.316 | -0.5922 | Yes | ||
| 120 | CGGBP1 | 4810053 | 17107 | -0.334 | -0.5838 | Yes | ||
| 121 | WASF1 | 2060484 | 17144 | -0.342 | -0.5739 | Yes | ||
| 122 | YTHDF2 | 4060332 4610279 | 17165 | -0.348 | -0.5630 | Yes | ||
| 123 | BCL2L11 | 780044 4200601 | 17172 | -0.349 | -0.5512 | Yes | ||
| 124 | RANBP10 | 2340184 | 17379 | -0.413 | -0.5481 | Yes | ||
| 125 | TRIM39 | 6550441 | 17476 | -0.447 | -0.5378 | Yes | ||
| 126 | EIF4G1 | 4070446 | 17543 | -0.470 | -0.5251 | Yes | ||
| 127 | SOX4 | 2260091 | 17766 | -0.562 | -0.5177 | Yes | ||
| 128 | KLF13 | 4670563 | 17786 | -0.570 | -0.4990 | Yes | ||
| 129 | ELOVL5 | 3800170 | 17844 | -0.598 | -0.4813 | Yes | ||
| 130 | AHCYL1 | 540609 | 17942 | -0.646 | -0.4642 | Yes | ||
| 131 | PDAP1 | 1410735 | 18081 | -0.739 | -0.4461 | Yes | ||
| 132 | PIK3R3 | 1770333 | 18093 | -0.744 | -0.4209 | Yes | ||
| 133 | DDX3X | 2190020 | 18098 | -0.751 | -0.3952 | Yes | ||
| 134 | FOXP1 | 6510433 | 18175 | -0.810 | -0.3712 | Yes | ||
| 135 | MAPK6 | 6760520 | 18240 | -0.875 | -0.3444 | Yes | ||
| 136 | KCNF1 | 2650687 | 18397 | -1.092 | -0.3151 | Yes | ||
| 137 | ABI1 | 2850152 | 18400 | -1.099 | -0.2772 | Yes | ||
| 138 | SFRS6 | 60224 | 18439 | -1.185 | -0.2382 | Yes | ||
| 139 | ILF3 | 940722 3190647 6520110 | 18576 | -2.079 | -0.1736 | Yes | ||
| 140 | CUTL1 | 1570113 1770091 1850273 2100286 5340605 | 18597 | -2.519 | -0.0875 | Yes | ||
| 141 | HNRPH1 | 1170086 3140546 3290471 6110184 6110373 | 18599 | -2.557 | 0.0009 | Yes |

