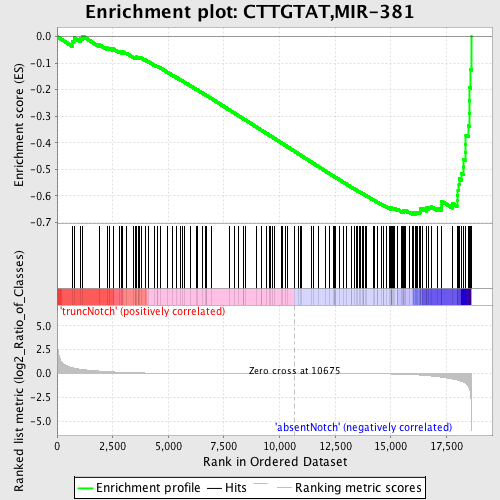
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #truncNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#truncNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | CTTGTAT,MIR-381 |
| Enrichment Score (ES) | -0.66957915 |
| Normalized Enrichment Score (NES) | -1.7136847 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.006643028 |
| FWER p-Value | 0.034 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GFRA1 | 3610152 | 679 | 0.604 | -0.0178 | No | ||
| 2 | DDX6 | 2630025 6770408 | 761 | 0.565 | -0.0044 | No | ||
| 3 | AGPAT3 | 60152 | 1053 | 0.444 | -0.0063 | No | ||
| 4 | CHD4 | 5420059 6130338 6380717 | 1150 | 0.415 | 0.0016 | No | ||
| 5 | PMP22 | 4010239 6550072 | 1893 | 0.259 | -0.0305 | No | ||
| 6 | ZMYND11 | 630181 2570019 4850138 5360040 | 2246 | 0.211 | -0.0429 | No | ||
| 7 | DHX40 | 360672 | 2371 | 0.191 | -0.0436 | No | ||
| 8 | RBM16 | 6290286 6400181 | 2515 | 0.171 | -0.0460 | No | ||
| 9 | BAT2 | 6650435 | 2787 | 0.135 | -0.0564 | No | ||
| 10 | INSIG1 | 1500332 3450458 4540301 | 2896 | 0.125 | -0.0584 | No | ||
| 11 | TMEM16D | 2650324 | 2926 | 0.121 | -0.0561 | No | ||
| 12 | ACTG1 | 540215 | 3102 | 0.104 | -0.0623 | No | ||
| 13 | CRSP2 | 1940059 | 3424 | 0.082 | -0.0771 | No | ||
| 14 | DLC1 | 1090632 6450594 | 3530 | 0.076 | -0.0804 | No | ||
| 15 | SRRM1 | 6110025 | 3538 | 0.076 | -0.0784 | No | ||
| 16 | NIN | 3450576 5080048 | 3547 | 0.075 | -0.0765 | No | ||
| 17 | PAPOLA | 4200286 4850112 | 3640 | 0.070 | -0.0793 | No | ||
| 18 | EIF3S10 | 1190079 3840176 | 3676 | 0.069 | -0.0790 | No | ||
| 19 | UBN1 | 5900195 | 3717 | 0.067 | -0.0790 | No | ||
| 20 | SIPA1L2 | 4850731 | 3783 | 0.064 | -0.0806 | No | ||
| 21 | UBE2E2 | 610053 | 3958 | 0.058 | -0.0882 | No | ||
| 22 | BCL11A | 6860369 | 4121 | 0.052 | -0.0953 | No | ||
| 23 | ROD1 | 2060324 3140332 | 4395 | 0.045 | -0.1087 | No | ||
| 24 | TRIM63 | 1740164 | 4490 | 0.042 | -0.1124 | No | ||
| 25 | ETS1 | 5270278 6450717 6620465 | 4503 | 0.042 | -0.1118 | No | ||
| 26 | COL3A1 | 6420273 6620044 | 4641 | 0.039 | -0.1180 | No | ||
| 27 | ST8SIA2 | 2680082 | 4974 | 0.032 | -0.1349 | No | ||
| 28 | VSNL1 | 3120017 | 5204 | 0.028 | -0.1465 | No | ||
| 29 | WNT5A | 840685 3120152 | 5387 | 0.025 | -0.1555 | No | ||
| 30 | HNRPR | 2320440 2900601 4610008 | 5524 | 0.024 | -0.1621 | No | ||
| 31 | GAD1 | 2360035 3140167 | 5619 | 0.023 | -0.1665 | No | ||
| 32 | CACNA1C | 1660128 3780300 3870092 6290403 6900301 | 5718 | 0.022 | -0.1711 | No | ||
| 33 | KIF1B | 1240494 2370139 4570270 6510102 | 6007 | 0.019 | -0.1861 | No | ||
| 34 | EPHA4 | 460750 | 6242 | 0.017 | -0.1983 | No | ||
| 35 | GRM8 | 4780082 | 6290 | 0.016 | -0.2003 | No | ||
| 36 | RAB11FIP2 | 4570040 | 6518 | 0.015 | -0.2121 | No | ||
| 37 | PDGFC | 4200133 | 6677 | 0.014 | -0.2203 | No | ||
| 38 | NAP1L5 | 360576 | 6698 | 0.013 | -0.2209 | No | ||
| 39 | CLCF1 | 4920176 6550750 | 6730 | 0.013 | -0.2222 | No | ||
| 40 | EPHA3 | 2190497 5080059 | 6923 | 0.012 | -0.2322 | No | ||
| 41 | FLRT3 | 70685 2760497 6040519 | 7758 | 0.008 | -0.2771 | No | ||
| 42 | SEMA6D | 4050324 5860138 6350307 | 7954 | 0.008 | -0.2874 | No | ||
| 43 | SHANK2 | 1500452 | 8171 | 0.007 | -0.2989 | No | ||
| 44 | ACVR2A | 6110647 | 8395 | 0.006 | -0.3108 | No | ||
| 45 | RNF13 | 2370021 | 8397 | 0.006 | -0.3106 | No | ||
| 46 | MDS1 | 6400072 | 8468 | 0.006 | -0.3142 | No | ||
| 47 | NFIA | 2760129 5860278 | 8971 | 0.004 | -0.3413 | No | ||
| 48 | STX12 | 610451 | 9193 | 0.004 | -0.3531 | No | ||
| 49 | RNF2 | 2340497 | 9204 | 0.004 | -0.3535 | No | ||
| 50 | MTSS1 | 780435 2370114 | 9421 | 0.003 | -0.3651 | No | ||
| 51 | RALGPS1 | 3440129 6400746 | 9547 | 0.003 | -0.3718 | No | ||
| 52 | ABCA1 | 6290156 | 9595 | 0.003 | -0.3743 | No | ||
| 53 | PNRC1 | 360025 | 9660 | 0.003 | -0.3777 | No | ||
| 54 | PARD6B | 4120600 | 9773 | 0.002 | -0.3837 | No | ||
| 55 | TBC1D15 | 870673 | 10078 | 0.001 | -0.4001 | No | ||
| 56 | NR5A2 | 1690180 6900050 | 10144 | 0.001 | -0.4036 | No | ||
| 57 | ZFPM2 | 460072 | 10273 | 0.001 | -0.4105 | No | ||
| 58 | RHOQ | 520161 | 10361 | 0.001 | -0.4151 | No | ||
| 59 | STAG1 | 1190300 1400722 4590100 | 10654 | 0.000 | -0.4310 | No | ||
| 60 | NRXN1 | 460408 5340270 6370066 | 10842 | -0.000 | -0.4411 | No | ||
| 61 | BIRC6 | 70148 | 10934 | -0.001 | -0.4460 | No | ||
| 62 | BTBD7 | 730288 | 10997 | -0.001 | -0.4493 | No | ||
| 63 | KLF12 | 1660095 4810288 5340546 6520286 | 11455 | -0.002 | -0.4740 | No | ||
| 64 | CNOT7 | 2450338 5720397 | 11531 | -0.002 | -0.4780 | No | ||
| 65 | GAS2 | 1340082 | 11751 | -0.003 | -0.4897 | No | ||
| 66 | MITF | 380056 | 12085 | -0.004 | -0.5077 | No | ||
| 67 | CCNT2 | 2100390 4150563 | 12259 | -0.005 | -0.5169 | No | ||
| 68 | FOXF2 | 1240091 | 12417 | -0.005 | -0.5252 | No | ||
| 69 | BHLHB5 | 6510520 | 12474 | -0.005 | -0.5281 | No | ||
| 70 | SYNCRIP | 1690195 3140113 4670279 | 12489 | -0.005 | -0.5287 | No | ||
| 71 | LRRC4 | 3850041 | 12521 | -0.006 | -0.5302 | No | ||
| 72 | JPH1 | 610739 | 12523 | -0.006 | -0.5301 | No | ||
| 73 | HNRPH2 | 2630040 5720671 | 12706 | -0.006 | -0.5397 | No | ||
| 74 | ZCCHC11 | 3390731 5360039 | 12876 | -0.007 | -0.5486 | No | ||
| 75 | ELOVL7 | 1400711 | 12990 | -0.008 | -0.5545 | No | ||
| 76 | HBP1 | 2510102 3130010 4210619 | 13215 | -0.009 | -0.5664 | No | ||
| 77 | RPL35A | 2370706 | 13229 | -0.009 | -0.5668 | No | ||
| 78 | SEMA3C | 7000020 | 13374 | -0.010 | -0.5743 | No | ||
| 79 | QKI | 5220093 6130044 | 13435 | -0.010 | -0.5772 | No | ||
| 80 | ACSL3 | 3140195 | 13498 | -0.011 | -0.5802 | No | ||
| 81 | SLC24A3 | 5080707 | 13579 | -0.011 | -0.5842 | No | ||
| 82 | AZIN1 | 3450070 | 13651 | -0.012 | -0.5877 | No | ||
| 83 | UBE3A | 1240152 2690438 5860609 | 13717 | -0.013 | -0.5908 | No | ||
| 84 | GRM1 | 5890324 | 13759 | -0.013 | -0.5926 | No | ||
| 85 | PBEF1 | 1090278 | 13761 | -0.013 | -0.5923 | No | ||
| 86 | B3GNT5 | 4780528 | 13862 | -0.014 | -0.5972 | No | ||
| 87 | RPS6KB1 | 1450427 5080110 6200563 | 13918 | -0.015 | -0.5997 | No | ||
| 88 | THRAP1 | 2470121 | 14234 | -0.020 | -0.6162 | No | ||
| 89 | SSH2 | 360056 | 14252 | -0.020 | -0.6165 | No | ||
| 90 | PURG | 1190593 | 14393 | -0.023 | -0.6233 | No | ||
| 91 | ATRN | 870735 3360390 | 14412 | -0.023 | -0.6236 | No | ||
| 92 | OSBPL3 | 3170164 4210196 | 14565 | -0.027 | -0.6310 | No | ||
| 93 | UBE4A | 6100520 | 14649 | -0.030 | -0.6345 | No | ||
| 94 | ARID4B | 730754 6660026 | 14657 | -0.030 | -0.6340 | No | ||
| 95 | XPO7 | 2630215 6760014 | 14787 | -0.034 | -0.6399 | No | ||
| 96 | CEBPA | 5690519 | 14947 | -0.041 | -0.6472 | No | ||
| 97 | CSTF3 | 3850156 5570458 | 14998 | -0.044 | -0.6485 | No | ||
| 98 | ASXL2 | 130670 | 15010 | -0.044 | -0.6478 | No | ||
| 99 | NLK | 2030010 2450041 | 15015 | -0.044 | -0.6466 | No | ||
| 100 | ROCK2 | 1780035 5720025 | 15017 | -0.044 | -0.6452 | No | ||
| 101 | R3HDM1 | 940685 3290161 | 15031 | -0.045 | -0.6445 | No | ||
| 102 | WEE1 | 3390070 | 15079 | -0.047 | -0.6456 | No | ||
| 103 | KLF4 | 1850022 3830239 5570750 | 15123 | -0.050 | -0.6464 | No | ||
| 104 | LPHN1 | 1400113 3130301 | 15179 | -0.053 | -0.6477 | No | ||
| 105 | PAPOLG | 2470400 | 15279 | -0.058 | -0.6512 | No | ||
| 106 | JAG2 | 1500341 | 15316 | -0.060 | -0.6513 | No | ||
| 107 | RNF111 | 2970072 3140112 5130647 | 15499 | -0.072 | -0.6589 | No | ||
| 108 | NDUFAB1 | 1980091 | 15533 | -0.074 | -0.6583 | No | ||
| 109 | LZTS2 | 2260440 | 15578 | -0.078 | -0.6583 | No | ||
| 110 | PHF15 | 6550066 6860673 | 15583 | -0.078 | -0.6560 | No | ||
| 111 | THRAP2 | 4920600 | 15592 | -0.079 | -0.6540 | No | ||
| 112 | SNRK | 630021 2030731 6350017 | 15655 | -0.083 | -0.6547 | No | ||
| 113 | KCTD15 | 2810735 | 15818 | -0.098 | -0.6604 | No | ||
| 114 | ITCH | 1410731 5700022 | 15988 | -0.116 | -0.6659 | Yes | ||
| 115 | HIPK1 | 110193 | 16015 | -0.118 | -0.6636 | Yes | ||
| 116 | KPNA3 | 7040088 | 16100 | -0.126 | -0.6642 | Yes | ||
| 117 | RPS6KA3 | 1980707 | 16162 | -0.134 | -0.6633 | Yes | ||
| 118 | ZFAND3 | 2760504 | 16199 | -0.139 | -0.6609 | Yes | ||
| 119 | GORASP2 | 2970059 4060309 7040706 | 16298 | -0.154 | -0.6614 | Yes | ||
| 120 | KCTD3 | 4120538 | 16333 | -0.159 | -0.6582 | Yes | ||
| 121 | NFKBIA | 1570152 | 16341 | -0.160 | -0.6536 | Yes | ||
| 122 | MBNL2 | 3450707 | 16344 | -0.161 | -0.6486 | Yes | ||
| 123 | UBR1 | 3800132 | 16423 | -0.174 | -0.6474 | Yes | ||
| 124 | ANKRD50 | 870543 | 16605 | -0.207 | -0.6507 | Yes | ||
| 125 | RNF144 | 4210332 | 16612 | -0.209 | -0.6444 | Yes | ||
| 126 | YTHDF1 | 4070739 | 16710 | -0.232 | -0.6424 | Yes | ||
| 127 | PHF2 | 4060300 | 16806 | -0.255 | -0.6395 | Yes | ||
| 128 | CGGBP1 | 4810053 | 17107 | -0.334 | -0.6453 | Yes | ||
| 129 | KHDRBS1 | 1240403 6040040 | 17265 | -0.373 | -0.6421 | Yes | ||
| 130 | SLC6A8 | 3940341 | 17283 | -0.377 | -0.6312 | Yes | ||
| 131 | SLC38A2 | 1470242 3800026 | 17287 | -0.378 | -0.6195 | Yes | ||
| 132 | SOX4 | 2260091 | 17766 | -0.562 | -0.6277 | Yes | ||
| 133 | BRD7 | 2120068 6200746 | 17977 | -0.664 | -0.6182 | Yes | ||
| 134 | ZFYVE20 | 6420400 | 17985 | -0.671 | -0.5975 | Yes | ||
| 135 | EIF1 | 5080100 | 18018 | -0.690 | -0.5775 | Yes | ||
| 136 | CXCR4 | 4590519 | 18063 | -0.720 | -0.5573 | Yes | ||
| 137 | PDAP1 | 1410735 | 18081 | -0.739 | -0.5350 | Yes | ||
| 138 | ETF1 | 6770075 | 18177 | -0.816 | -0.5145 | Yes | ||
| 139 | RAP2C | 1690132 | 18260 | -0.898 | -0.4907 | Yes | ||
| 140 | BCL11B | 2680673 | 18267 | -0.908 | -0.4625 | Yes | ||
| 141 | APRIN | 2810050 | 18339 | -0.987 | -0.4353 | Yes | ||
| 142 | EIF4G3 | 670707 | 18369 | -1.035 | -0.4044 | Yes | ||
| 143 | APPBP2 | 5130215 | 18376 | -1.048 | -0.3718 | Yes | ||
| 144 | SATB1 | 5670154 | 18479 | -1.341 | -0.3352 | Yes | ||
| 145 | EIF4G2 | 3800575 6860184 | 18522 | -1.497 | -0.2904 | Yes | ||
| 146 | PSIP1 | 1780082 3190435 5050594 | 18531 | -1.551 | -0.2421 | Yes | ||
| 147 | SLMAP | 3120093 4760128 5390528 | 18542 | -1.608 | -0.1921 | Yes | ||
| 148 | GJA1 | 5220731 | 18585 | -2.218 | -0.1247 | Yes | ||
| 149 | GTF2I | 2470750 3870408 3830369 3990082 4610138 | 18614 | -4.021 | 0.0001 | Yes |

