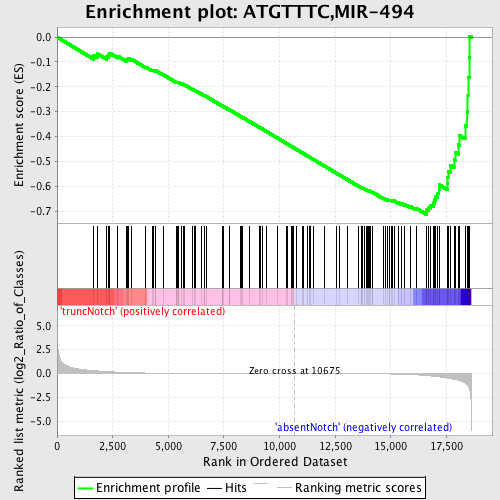
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #truncNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#truncNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | ATGTTTC,MIR-494 |
| Enrichment Score (ES) | -0.7137695 |
| Normalized Enrichment Score (NES) | -1.7871983 |
| Nominal p-value | 0.0 |
| FDR q-value | 4.1913748E-4 |
| FWER p-Value | 0.0010 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CAMK2D | 2370685 | 1639 | 0.305 | -0.0723 | No | ||
| 2 | CLTA | 450010 | 1800 | 0.276 | -0.0662 | No | ||
| 3 | NUTF2 | 1410471 5720050 | 2224 | 0.214 | -0.0777 | No | ||
| 4 | HIP2 | 2810095 3990369 4120301 | 2310 | 0.199 | -0.0717 | No | ||
| 5 | SON | 3120148 5860239 7040403 | 2364 | 0.192 | -0.0644 | No | ||
| 6 | DNAJA2 | 2630253 6860358 | 2730 | 0.142 | -0.0766 | No | ||
| 7 | HAS2 | 5360181 | 3114 | 0.103 | -0.0918 | No | ||
| 8 | HTRA1 | 3450601 | 3154 | 0.100 | -0.0886 | No | ||
| 9 | SFRS8 | 5130162 5700164 | 3206 | 0.096 | -0.0862 | No | ||
| 10 | EGR3 | 6940128 | 3356 | 0.086 | -0.0897 | No | ||
| 11 | ING3 | 110438 2060390 | 3989 | 0.056 | -0.1208 | No | ||
| 12 | BMPR2 | 5340066 6960328 | 4265 | 0.048 | -0.1331 | No | ||
| 13 | NEUROD1 | 3060619 | 4321 | 0.047 | -0.1336 | No | ||
| 14 | RB1CC1 | 510494 7100072 | 4412 | 0.044 | -0.1361 | No | ||
| 15 | PCOLCE | 4210433 | 4436 | 0.044 | -0.1350 | No | ||
| 16 | PTER | 3450685 | 4781 | 0.036 | -0.1517 | No | ||
| 17 | ELF2 | 1690014 2480132 3060180 3190504 | 5369 | 0.026 | -0.1820 | No | ||
| 18 | POU3F2 | 6620484 | 5382 | 0.026 | -0.1813 | No | ||
| 19 | DCBLD2 | 5890333 | 5432 | 0.025 | -0.1827 | No | ||
| 20 | ZZZ3 | 780133 2320239 2640452 5080706 6550341 | 5446 | 0.025 | -0.1820 | No | ||
| 21 | NHLH2 | 6200021 | 5458 | 0.025 | -0.1813 | No | ||
| 22 | CLK1 | 1780551 2100102 2340347 2480347 | 5604 | 0.023 | -0.1879 | No | ||
| 23 | ARNT | 1170672 5670711 | 5678 | 0.022 | -0.1907 | No | ||
| 24 | CACNA1C | 1660128 3780300 3870092 6290403 6900301 | 5718 | 0.022 | -0.1916 | No | ||
| 25 | LIF | 1410373 2060129 3990338 | 6068 | 0.018 | -0.2095 | No | ||
| 26 | ATRX | 2340039 3610132 4210373 | 6152 | 0.018 | -0.2131 | No | ||
| 27 | HOXB5 | 2900538 | 6224 | 0.017 | -0.2160 | No | ||
| 28 | EIF5A2 | 7040139 | 6468 | 0.015 | -0.2283 | No | ||
| 29 | GRIK2 | 4610164 | 6602 | 0.014 | -0.2348 | No | ||
| 30 | LRP1B | 1450193 | 6605 | 0.014 | -0.2341 | No | ||
| 31 | DSCAML1 | 1850403 5550079 | 6699 | 0.013 | -0.2384 | No | ||
| 32 | ARHGEF17 | 2060008 | 7441 | 0.010 | -0.2780 | No | ||
| 33 | PHOX2B | 5270075 | 7442 | 0.010 | -0.2775 | No | ||
| 34 | PITX2 | 870537 2690139 | 7494 | 0.009 | -0.2797 | No | ||
| 35 | CNIH | 2190484 5080609 | 7738 | 0.008 | -0.2924 | No | ||
| 36 | DPYSL3 | 1580594 | 7759 | 0.008 | -0.2930 | No | ||
| 37 | DMD | 1740041 3990332 | 8245 | 0.007 | -0.3189 | No | ||
| 38 | SLC26A3 | 380750 | 8304 | 0.006 | -0.3217 | No | ||
| 39 | ARHGEF12 | 3990195 | 8327 | 0.006 | -0.3225 | No | ||
| 40 | TAC1 | 7000195 380706 | 8648 | 0.005 | -0.3395 | No | ||
| 41 | LRRC19 | 2680025 | 9089 | 0.004 | -0.3631 | No | ||
| 42 | EIF2C3 | 2450348 3130332 6980487 | 9123 | 0.004 | -0.3647 | No | ||
| 43 | LMO1 | 3170528 | 9249 | 0.004 | -0.3712 | No | ||
| 44 | MYLIP | 50717 4050672 | 9423 | 0.003 | -0.3804 | No | ||
| 45 | NF2 | 4150735 6450139 | 9906 | 0.002 | -0.4063 | No | ||
| 46 | EIF2C4 | 2940577 5900563 | 10315 | 0.001 | -0.4283 | No | ||
| 47 | ELOVL3 | 6860050 | 10376 | 0.001 | -0.4315 | No | ||
| 48 | SP4 | 3850176 | 10522 | 0.000 | -0.4394 | No | ||
| 49 | ZIC2 | 1410408 | 10560 | 0.000 | -0.4413 | No | ||
| 50 | PHF8 | 70722 | 10643 | 0.000 | -0.4458 | No | ||
| 51 | SLC25A16 | 4200341 | 10780 | -0.000 | -0.4531 | No | ||
| 52 | EBF3 | 4200129 | 11023 | -0.001 | -0.4661 | No | ||
| 53 | OSBPL6 | 3800072 | 11082 | -0.001 | -0.4692 | No | ||
| 54 | ADAM10 | 3780156 | 11267 | -0.002 | -0.4791 | No | ||
| 55 | PRICKLE2 | 780408 | 11362 | -0.002 | -0.4841 | No | ||
| 56 | GALNT7 | 3990280 | 11394 | -0.002 | -0.4856 | No | ||
| 57 | LMX1A | 2810239 5690324 | 11521 | -0.002 | -0.4923 | No | ||
| 58 | GPRC6A | 4560576 | 12011 | -0.004 | -0.5186 | No | ||
| 59 | ARHGAP5 | 2510619 3360035 | 12565 | -0.006 | -0.5481 | No | ||
| 60 | PRPF4B | 940181 2570300 | 12696 | -0.006 | -0.5548 | No | ||
| 61 | PCSK1 | 2350148 | 13031 | -0.008 | -0.5724 | No | ||
| 62 | SSX2IP | 4610176 | 13554 | -0.011 | -0.6001 | No | ||
| 63 | UBP1 | 6020707 | 13665 | -0.012 | -0.6054 | No | ||
| 64 | GABRA5 | 5570215 | 13688 | -0.012 | -0.6059 | No | ||
| 65 | PTPN12 | 2030309 6020725 6130746 6290609 | 13734 | -0.013 | -0.6076 | No | ||
| 66 | API5 | 540593 1740114 6980692 | 13833 | -0.014 | -0.6122 | No | ||
| 67 | MESDC1 | 2030019 | 13914 | -0.015 | -0.6157 | No | ||
| 68 | CANX | 4760402 | 13932 | -0.015 | -0.6158 | No | ||
| 69 | ATF3 | 1940546 | 13979 | -0.016 | -0.6175 | No | ||
| 70 | STAC2 | 7100022 | 14024 | -0.016 | -0.6190 | No | ||
| 71 | RNF20 | 3060717 | 14078 | -0.017 | -0.6210 | No | ||
| 72 | DMWD | 770019 1690047 | 14170 | -0.018 | -0.6249 | No | ||
| 73 | PPARGC1A | 4670040 | 14182 | -0.019 | -0.6245 | No | ||
| 74 | ARID4B | 730754 6660026 | 14657 | -0.030 | -0.6485 | No | ||
| 75 | SLITRK2 | 2640020 | 14757 | -0.033 | -0.6521 | No | ||
| 76 | DBN1 | 7100537 | 14835 | -0.036 | -0.6543 | No | ||
| 77 | FGFR2 | 4670601 5570402 6940377 | 14922 | -0.040 | -0.6569 | No | ||
| 78 | WDR44 | 130195 1050609 1570685 | 14950 | -0.041 | -0.6561 | No | ||
| 79 | LASS6 | 5890576 6520138 | 15009 | -0.044 | -0.6569 | No | ||
| 80 | TRPS1 | 870100 | 15096 | -0.049 | -0.6590 | No | ||
| 81 | CRKRS | 510537 2510551 | 15143 | -0.051 | -0.6588 | No | ||
| 82 | CHD9 | 430037 2570129 5420398 | 15355 | -0.063 | -0.6668 | No | ||
| 83 | FUT8 | 1340068 2340056 | 15476 | -0.070 | -0.6696 | No | ||
| 84 | TACC2 | 580092 1400364 2650497 2690315 5360338 | 15619 | -0.080 | -0.6729 | No | ||
| 85 | CUL4A | 1170088 2320008 2470278 | 15864 | -0.101 | -0.6808 | No | ||
| 86 | FOXJ2 | 2450441 | 16136 | -0.130 | -0.6885 | No | ||
| 87 | ANKRD50 | 870543 | 16605 | -0.207 | -0.7028 | Yes | ||
| 88 | ARNTL | 3170463 | 16623 | -0.211 | -0.6924 | Yes | ||
| 89 | SFRS7 | 2760408 | 16712 | -0.232 | -0.6848 | Yes | ||
| 90 | LANCL2 | 460047 | 16793 | -0.252 | -0.6757 | Yes | ||
| 91 | PURB | 5360138 | 16916 | -0.280 | -0.6674 | Yes | ||
| 92 | BHLHB2 | 7040603 | 16960 | -0.290 | -0.6543 | Yes | ||
| 93 | ATP10A | 4780050 | 17021 | -0.309 | -0.6411 | Yes | ||
| 94 | CGGBP1 | 4810053 | 17107 | -0.334 | -0.6279 | Yes | ||
| 95 | YTHDF2 | 4060332 4610279 | 17165 | -0.348 | -0.6124 | Yes | ||
| 96 | OGT | 2360131 4610333 | 17176 | -0.350 | -0.5943 | Yes | ||
| 97 | ZRANB3 | 3990364 | 17531 | -0.465 | -0.5887 | Yes | ||
| 98 | CUGBP2 | 6180121 6840139 | 17550 | -0.471 | -0.5645 | Yes | ||
| 99 | MMD | 3850239 | 17572 | -0.486 | -0.5398 | Yes | ||
| 100 | SMARCE1 | 4920465 4390398 | 17698 | -0.538 | -0.5178 | Yes | ||
| 101 | ELOVL5 | 3800170 | 17844 | -0.598 | -0.4938 | Yes | ||
| 102 | IKBKAP | 1740746 3120364 | 17891 | -0.622 | -0.4632 | Yes | ||
| 103 | ACACA | 2490612 2680369 | 18033 | -0.701 | -0.4334 | Yes | ||
| 104 | MORF4L2 | 6450133 | 18097 | -0.750 | -0.3969 | Yes | ||
| 105 | H3F3A | 2900086 | 18341 | -0.991 | -0.3572 | Yes | ||
| 106 | RAP1B | 3710138 | 18424 | -1.148 | -0.3005 | Yes | ||
| 107 | EIF5 | 6380750 | 18467 | -1.296 | -0.2337 | Yes | ||
| 108 | H3F3B | 1410300 | 18492 | -1.379 | -0.1615 | Yes | ||
| 109 | HMGCS1 | 6620452 | 18530 | -1.546 | -0.0812 | Yes | ||
| 110 | CCND2 | 5340167 | 18543 | -1.609 | 0.0039 | Yes |

