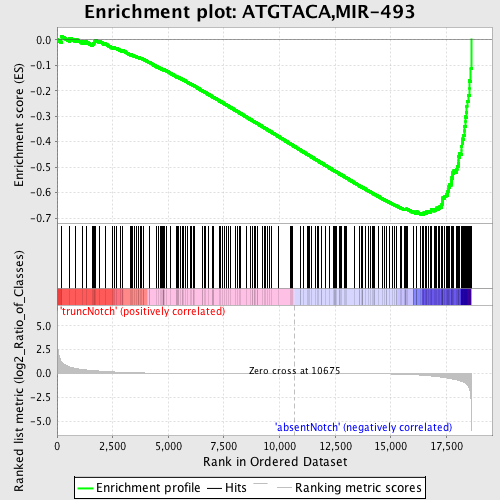
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #truncNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#truncNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | ATGTACA,MIR-493 |
| Enrichment Score (ES) | -0.68551385 |
| Normalized Enrichment Score (NES) | -1.8416406 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | UBE2Q1 | 610348 2350114 4210717 | 186 | 1.259 | 0.0142 | No | ||
| 2 | E2F6 | 3120133 3850332 4070324 | 570 | 0.680 | 0.0065 | No | ||
| 3 | CARM1 | 7040292 | 829 | 0.527 | 0.0027 | No | ||
| 4 | OTUB1 | 610551 | 1160 | 0.412 | -0.0073 | No | ||
| 5 | VAMP2 | 2100100 | 1312 | 0.372 | -0.0083 | No | ||
| 6 | ATP5A1 | 1990722 | 1603 | 0.311 | -0.0181 | No | ||
| 7 | FBS1 | 2570520 | 1643 | 0.305 | -0.0143 | No | ||
| 8 | CHKB | 770129 | 1663 | 0.300 | -0.0095 | No | ||
| 9 | CRSP7 | 4230408 | 1694 | 0.294 | -0.0055 | No | ||
| 10 | IRF7 | 1570605 | 1741 | 0.286 | -0.0025 | No | ||
| 11 | PMP22 | 4010239 6550072 | 1893 | 0.259 | -0.0057 | No | ||
| 12 | SNRP70 | 4200181 | 2168 | 0.220 | -0.0163 | No | ||
| 13 | EFNA3 | 6370390 | 2468 | 0.178 | -0.0291 | No | ||
| 14 | KLF3 | 5130438 | 2560 | 0.167 | -0.0308 | No | ||
| 15 | BAZ2A | 730184 | 2653 | 0.153 | -0.0329 | No | ||
| 16 | MAST1 | 6940215 | 2851 | 0.129 | -0.0411 | No | ||
| 17 | TMEM16D | 2650324 | 2926 | 0.121 | -0.0428 | No | ||
| 18 | USP21 | 3060446 | 2938 | 0.119 | -0.0411 | No | ||
| 19 | SH2D3C | 3060014 | 3318 | 0.088 | -0.0600 | No | ||
| 20 | DOCK9 | 5080037 | 3323 | 0.088 | -0.0585 | No | ||
| 21 | HNRPA0 | 2680048 | 3393 | 0.084 | -0.0606 | No | ||
| 22 | AXIN1 | 3360358 6940451 | 3479 | 0.079 | -0.0637 | No | ||
| 23 | HIC2 | 4010433 | 3558 | 0.075 | -0.0665 | No | ||
| 24 | BCL2 | 730132 1570736 2470138 3800044 4810037 5690068 5860504 6650164 | 3639 | 0.070 | -0.0695 | No | ||
| 25 | ATAD2 | 870242 2260687 | 3678 | 0.069 | -0.0702 | No | ||
| 26 | SPRYD3 | 50403 | 3764 | 0.065 | -0.0736 | No | ||
| 27 | SIPA1L2 | 4850731 | 3783 | 0.064 | -0.0733 | No | ||
| 28 | ZMYND19 | 1570706 6860551 | 3896 | 0.059 | -0.0783 | No | ||
| 29 | KPNA4 | 510369 | 3903 | 0.059 | -0.0775 | No | ||
| 30 | GABRG2 | 2350402 4210204 6130279 6550037 | 4156 | 0.051 | -0.0902 | No | ||
| 31 | UNC5A | 2120528 3060324 | 4171 | 0.051 | -0.0900 | No | ||
| 32 | CDH2 | 520435 2450451 2760025 6650477 | 4488 | 0.042 | -0.1063 | No | ||
| 33 | RNF38 | 1940746 3140133 3840601 | 4560 | 0.040 | -0.1094 | No | ||
| 34 | TBC1D4 | 6420397 | 4649 | 0.038 | -0.1134 | No | ||
| 35 | SDC2 | 1980168 | 4673 | 0.038 | -0.1140 | No | ||
| 36 | HIVEP2 | 6520538 | 4750 | 0.036 | -0.1174 | No | ||
| 37 | SPEN | 2060041 | 4776 | 0.036 | -0.1180 | No | ||
| 38 | SALL1 | 5420020 7050195 | 4792 | 0.036 | -0.1182 | No | ||
| 39 | RPS6KA5 | 2120563 7040546 | 4797 | 0.035 | -0.1177 | No | ||
| 40 | PPP2R5C | 3310121 6650672 | 4802 | 0.035 | -0.1172 | No | ||
| 41 | PBX3 | 1300424 3710577 6180575 | 4815 | 0.035 | -0.1172 | No | ||
| 42 | SNAI1 | 6590253 | 4903 | 0.034 | -0.1213 | No | ||
| 43 | SPRED2 | 510138 540195 | 5117 | 0.030 | -0.1323 | No | ||
| 44 | FGF13 | 630575 1570440 5360121 | 5379 | 0.026 | -0.1460 | No | ||
| 45 | WNT5A | 840685 3120152 | 5387 | 0.025 | -0.1459 | No | ||
| 46 | HS3ST3B1 | 380040 | 5417 | 0.025 | -0.1470 | No | ||
| 47 | COBLL1 | 4230056 6370494 | 5423 | 0.025 | -0.1468 | No | ||
| 48 | EPHA7 | 1190288 4010278 | 5441 | 0.025 | -0.1472 | No | ||
| 49 | SORCS3 | 5700309 | 5442 | 0.025 | -0.1467 | No | ||
| 50 | ENAH | 1690292 5700300 | 5557 | 0.024 | -0.1525 | No | ||
| 51 | GAD1 | 2360035 3140167 | 5619 | 0.023 | -0.1553 | No | ||
| 52 | SPAG8 | 4280446 | 5649 | 0.023 | -0.1565 | No | ||
| 53 | CRSP6 | 2940358 | 5674 | 0.022 | -0.1574 | No | ||
| 54 | AR | 6380167 | 5749 | 0.021 | -0.1610 | No | ||
| 55 | SEMA4C | 1980315 | 5881 | 0.020 | -0.1677 | No | ||
| 56 | KIF1B | 1240494 2370139 4570270 6510102 | 6007 | 0.019 | -0.1741 | No | ||
| 57 | FHOD3 | 1240541 | 6027 | 0.019 | -0.1748 | No | ||
| 58 | PCGF2 | 6370347 | 6128 | 0.018 | -0.1799 | No | ||
| 59 | NDST1 | 1500427 5340121 | 6133 | 0.018 | -0.1798 | No | ||
| 60 | VPS13D | 2350451 | 6158 | 0.017 | -0.1807 | No | ||
| 61 | TTN | 2320161 4670056 6550026 | 6532 | 0.015 | -0.2007 | No | ||
| 62 | TRAM2 | 3940112 | 6537 | 0.015 | -0.2007 | No | ||
| 63 | FZD4 | 1580520 | 6643 | 0.014 | -0.2061 | No | ||
| 64 | PDGFC | 4200133 | 6677 | 0.014 | -0.2076 | No | ||
| 65 | UNC50 | 4920037 | 6805 | 0.013 | -0.2143 | No | ||
| 66 | NXPH1 | 3870546 | 7000 | 0.012 | -0.2246 | No | ||
| 67 | SV2B | 3850717 | 7003 | 0.012 | -0.2245 | No | ||
| 68 | BASP1 | 5700044 | 7027 | 0.012 | -0.2255 | No | ||
| 69 | AK5 | 6290348 | 7039 | 0.011 | -0.2259 | No | ||
| 70 | POU4F1 | 2260315 | 7302 | 0.010 | -0.2399 | No | ||
| 71 | ZIC1 | 670113 | 7354 | 0.010 | -0.2425 | No | ||
| 72 | DKK2 | 3610433 | 7413 | 0.010 | -0.2455 | No | ||
| 73 | TMEFF1 | 2680100 | 7542 | 0.009 | -0.2523 | No | ||
| 74 | LRRTM3 | 4210021 | 7601 | 0.009 | -0.2553 | No | ||
| 75 | SLC25A26 | 4210577 | 7682 | 0.009 | -0.2594 | No | ||
| 76 | CDH11 | 1230133 2100180 4610110 | 7792 | 0.008 | -0.2652 | No | ||
| 77 | EIF2C1 | 6900551 | 8027 | 0.007 | -0.2778 | No | ||
| 78 | AFF3 | 3870102 | 8110 | 0.007 | -0.2821 | No | ||
| 79 | GABRA1 | 130372 | 8190 | 0.007 | -0.2863 | No | ||
| 80 | IL1A | 3610056 | 8221 | 0.007 | -0.2878 | No | ||
| 81 | BHLHB3 | 3450438 | 8523 | 0.006 | -0.3040 | No | ||
| 82 | PIK3R1 | 4730671 | 8693 | 0.005 | -0.3131 | No | ||
| 83 | WDFY3 | 3610041 4560333 4570273 | 8760 | 0.005 | -0.3166 | No | ||
| 84 | EYA1 | 1450278 5220390 | 8793 | 0.005 | -0.3182 | No | ||
| 85 | FOXO1A | 3120670 | 8886 | 0.005 | -0.3231 | No | ||
| 86 | GPC4 | 5910408 | 8888 | 0.005 | -0.3231 | No | ||
| 87 | UBE2V2 | 4210484 | 8908 | 0.005 | -0.3241 | No | ||
| 88 | SS18 | 1940440 2900072 6900129 | 8997 | 0.004 | -0.3288 | No | ||
| 89 | NRXN3 | 1400546 3360471 | 9006 | 0.004 | -0.3291 | No | ||
| 90 | ZBTB11 | 6520092 | 9247 | 0.004 | -0.3421 | No | ||
| 91 | SRPX | 580717 1980162 | 9310 | 0.003 | -0.3454 | No | ||
| 92 | CPNE8 | 7040039 | 9378 | 0.003 | -0.3490 | No | ||
| 93 | DACH1 | 2450593 5700592 | 9457 | 0.003 | -0.3531 | No | ||
| 94 | RALGPS1 | 3440129 6400746 | 9547 | 0.003 | -0.3579 | No | ||
| 95 | SPIRE2 | 450088 | 9628 | 0.003 | -0.3622 | No | ||
| 96 | PSD2 | 2060091 | 9945 | 0.002 | -0.3794 | No | ||
| 97 | NAB1 | 430739 1230114 3390736 6660056 | 10501 | 0.000 | -0.4095 | No | ||
| 98 | SP4 | 3850176 | 10522 | 0.000 | -0.4106 | No | ||
| 99 | ZIC2 | 1410408 | 10560 | 0.000 | -0.4126 | No | ||
| 100 | PUM2 | 4200441 5910446 | 10921 | -0.001 | -0.4322 | No | ||
| 101 | SERTAD2 | 2810673 6840450 | 10956 | -0.001 | -0.4340 | No | ||
| 102 | OSBPL6 | 3800072 | 11082 | -0.001 | -0.4408 | No | ||
| 103 | ADAM10 | 3780156 | 11267 | -0.002 | -0.4508 | No | ||
| 104 | TLE4 | 1450064 1500519 | 11289 | -0.002 | -0.4519 | No | ||
| 105 | PDZRN3 | 2340131 | 11351 | -0.002 | -0.4552 | No | ||
| 106 | PFN2 | 730040 5700026 | 11440 | -0.002 | -0.4599 | No | ||
| 107 | NLGN1 | 5670278 | 11625 | -0.003 | -0.4699 | No | ||
| 108 | GRM5 | 60528 | 11705 | -0.003 | -0.4741 | No | ||
| 109 | CUL5 | 450142 | 11747 | -0.003 | -0.4763 | No | ||
| 110 | KCNIP4 | 4120435 7050605 | 11867 | -0.003 | -0.4827 | No | ||
| 111 | RPE | 940114 | 11890 | -0.003 | -0.4838 | No | ||
| 112 | PCDH18 | 430010 | 12045 | -0.004 | -0.4921 | No | ||
| 113 | CCNT2 | 2100390 4150563 | 12259 | -0.005 | -0.5036 | No | ||
| 114 | DLL1 | 1770377 | 12429 | -0.005 | -0.5127 | No | ||
| 115 | IRX3 | 4200112 | 12460 | -0.005 | -0.5142 | No | ||
| 116 | BHLHB5 | 6510520 | 12474 | -0.005 | -0.5148 | No | ||
| 117 | JPH1 | 610739 | 12523 | -0.006 | -0.5173 | No | ||
| 118 | OVOL1 | 1690131 | 12526 | -0.006 | -0.5173 | No | ||
| 119 | ARHGAP5 | 2510619 3360035 | 12565 | -0.006 | -0.5193 | No | ||
| 120 | KCNMA1 | 1450402 | 12686 | -0.006 | -0.5257 | No | ||
| 121 | FIGN | 7000601 | 12712 | -0.006 | -0.5269 | No | ||
| 122 | BCOR | 1660452 3940053 | 12753 | -0.007 | -0.5290 | No | ||
| 123 | MEF2C | 670025 780338 | 12780 | -0.007 | -0.5302 | No | ||
| 124 | SNX9 | 3290592 | 12931 | -0.007 | -0.5383 | No | ||
| 125 | ESRRG | 4010181 | 12954 | -0.007 | -0.5393 | No | ||
| 126 | RSBN1 | 7000487 | 12985 | -0.008 | -0.5408 | No | ||
| 127 | SP3 | 3840338 | 13383 | -0.010 | -0.5622 | No | ||
| 128 | SOX21 | 1050110 4670041 6660338 | 13574 | -0.011 | -0.5723 | No | ||
| 129 | JUN | 840170 | 13604 | -0.011 | -0.5736 | No | ||
| 130 | GABRA5 | 5570215 | 13688 | -0.012 | -0.5779 | No | ||
| 131 | UBE3A | 1240152 2690438 5860609 | 13717 | -0.013 | -0.5792 | No | ||
| 132 | TJP1 | 6350184 | 13719 | -0.013 | -0.5790 | No | ||
| 133 | PTPN12 | 2030309 6020725 6130746 6290609 | 13734 | -0.013 | -0.5795 | No | ||
| 134 | RIMS3 | 3890563 | 13849 | -0.014 | -0.5855 | No | ||
| 135 | MARK1 | 450484 | 14010 | -0.016 | -0.5938 | No | ||
| 136 | CSNK1A1 | 2340427 | 14088 | -0.017 | -0.5977 | No | ||
| 137 | NLGN3 | 2650463 | 14173 | -0.018 | -0.6019 | No | ||
| 138 | THRAP1 | 2470121 | 14234 | -0.020 | -0.6048 | No | ||
| 139 | SSH2 | 360056 | 14252 | -0.020 | -0.6053 | No | ||
| 140 | PIP5K3 | 5360112 | 14260 | -0.020 | -0.6053 | No | ||
| 141 | DUSP16 | 2370082 2480450 | 14464 | -0.024 | -0.6159 | No | ||
| 142 | TNFSF11 | 6350204 | 14630 | -0.029 | -0.6243 | No | ||
| 143 | SVIL | 2450347 4050735 4070576 4120706 5390161 5860142 6520278 7100397 | 14699 | -0.032 | -0.6274 | No | ||
| 144 | XPO7 | 2630215 6760014 | 14787 | -0.034 | -0.6314 | No | ||
| 145 | GTF2IRD1 | 2030154 3990040 5080372 | 14797 | -0.035 | -0.6313 | No | ||
| 146 | CDH13 | 5130368 5340114 | 14920 | -0.040 | -0.6371 | No | ||
| 147 | TRPS1 | 870100 | 15096 | -0.049 | -0.6457 | No | ||
| 148 | PDE4D | 2470528 6660014 | 15170 | -0.052 | -0.6487 | No | ||
| 149 | DCUN1D1 | 2360332 | 15243 | -0.056 | -0.6515 | No | ||
| 150 | CITED2 | 5670114 5130088 | 15257 | -0.057 | -0.6511 | No | ||
| 151 | ARFGEF1 | 6760494 | 15432 | -0.068 | -0.6593 | No | ||
| 152 | CAPN3 | 1980451 6940047 | 15498 | -0.072 | -0.6614 | No | ||
| 153 | THRAP2 | 4920600 | 15592 | -0.079 | -0.6649 | No | ||
| 154 | GAB1 | 2970156 | 15640 | -0.082 | -0.6659 | No | ||
| 155 | ANKRD17 | 2120402 3850035 6400044 | 15674 | -0.085 | -0.6661 | No | ||
| 156 | MADD | 6100725 7000088 | 15675 | -0.085 | -0.6644 | No | ||
| 157 | MLLT10 | 1410288 3290142 | 15695 | -0.087 | -0.6638 | No | ||
| 158 | RBBP6 | 2320129 | 15752 | -0.092 | -0.6650 | No | ||
| 159 | HIPK1 | 110193 | 16015 | -0.118 | -0.6770 | No | ||
| 160 | RBM12 | 4570239 6040411 6660129 | 16029 | -0.120 | -0.6754 | No | ||
| 161 | CLASP1 | 6860279 | 16134 | -0.130 | -0.6785 | No | ||
| 162 | CDC14A | 4050132 | 16158 | -0.133 | -0.6772 | No | ||
| 163 | TTC7B | 4760725 | 16170 | -0.135 | -0.6752 | No | ||
| 164 | MBNL2 | 3450707 | 16344 | -0.161 | -0.6815 | No | ||
| 165 | ITSN2 | 780484 | 16419 | -0.173 | -0.6822 | Yes | ||
| 166 | EFTUD1 | 5360736 | 16478 | -0.185 | -0.6818 | Yes | ||
| 167 | HEMGN | 670411 | 16489 | -0.186 | -0.6787 | Yes | ||
| 168 | DNAJC13 | 4730091 | 16570 | -0.200 | -0.6792 | Yes | ||
| 169 | MAPK1 | 3190193 6200253 | 16598 | -0.206 | -0.6767 | Yes | ||
| 170 | ANKRD50 | 870543 | 16605 | -0.207 | -0.6730 | Yes | ||
| 171 | PHF12 | 870400 3130687 | 16672 | -0.222 | -0.6723 | Yes | ||
| 172 | VGLL4 | 6860463 | 16801 | -0.254 | -0.6744 | Yes | ||
| 173 | CDC73 | 1580132 | 16805 | -0.255 | -0.6696 | Yes | ||
| 174 | PHF2 | 4060300 | 16806 | -0.255 | -0.6647 | Yes | ||
| 175 | JARID1B | 4730494 | 16941 | -0.286 | -0.6664 | Yes | ||
| 176 | IRF2 | 5390044 | 17022 | -0.309 | -0.6648 | Yes | ||
| 177 | BCL7A | 4560095 | 17047 | -0.316 | -0.6600 | Yes | ||
| 178 | FBXO33 | 1510504 | 17154 | -0.345 | -0.6591 | Yes | ||
| 179 | EIF4ENIF1 | 2060341 | 17205 | -0.356 | -0.6549 | Yes | ||
| 180 | NCOA1 | 3610438 | 17281 | -0.377 | -0.6517 | Yes | ||
| 181 | SFRS3 | 770315 4230593 | 17293 | -0.381 | -0.6450 | Yes | ||
| 182 | GNB1 | 2120397 | 17307 | -0.386 | -0.6382 | Yes | ||
| 183 | LRP12 | 2370112 | 17326 | -0.395 | -0.6316 | Yes | ||
| 184 | PPP2R5D | 4010156 380408 5550112 5670162 | 17331 | -0.396 | -0.6242 | Yes | ||
| 185 | ZFX | 5900400 | 17341 | -0.399 | -0.6169 | Yes | ||
| 186 | PAIP1 | 3870576 | 17422 | -0.427 | -0.6130 | Yes | ||
| 187 | NMT1 | 5810152 | 17482 | -0.448 | -0.6076 | Yes | ||
| 188 | CUGBP2 | 6180121 6840139 | 17550 | -0.471 | -0.6021 | Yes | ||
| 189 | RYBP | 60746 | 17563 | -0.481 | -0.5935 | Yes | ||
| 190 | PDPK1 | 6650168 | 17599 | -0.496 | -0.5858 | Yes | ||
| 191 | UTX | 2900017 6770452 | 17610 | -0.499 | -0.5767 | Yes | ||
| 192 | LRIG1 | 1050136 | 17651 | -0.513 | -0.5690 | Yes | ||
| 193 | CREBBP | 5690035 7040050 | 17707 | -0.541 | -0.5615 | Yes | ||
| 194 | DHX36 | 2470465 | 17729 | -0.548 | -0.5521 | Yes | ||
| 195 | FMNL2 | 4060438 | 17742 | -0.554 | -0.5420 | Yes | ||
| 196 | NUDT4 | 6760689 | 17765 | -0.562 | -0.5324 | Yes | ||
| 197 | MTPN | 3940167 | 17784 | -0.569 | -0.5223 | Yes | ||
| 198 | CTDSPL | 4670546 | 17836 | -0.596 | -0.5136 | Yes | ||
| 199 | KBTBD2 | 2320014 | 17959 | -0.655 | -0.5076 | Yes | ||
| 200 | BRD7 | 2120068 6200746 | 17977 | -0.664 | -0.4957 | Yes | ||
| 201 | PPP1R10 | 110136 3170725 3610484 | 18027 | -0.699 | -0.4848 | Yes | ||
| 202 | ANTXR1 | 1990593 2850056 | 18045 | -0.708 | -0.4721 | Yes | ||
| 203 | SIN3A | 2190121 | 18047 | -0.708 | -0.4584 | Yes | ||
| 204 | DDX3X | 2190020 | 18098 | -0.751 | -0.4466 | Yes | ||
| 205 | SRPK2 | 6380341 | 18176 | -0.812 | -0.4351 | Yes | ||
| 206 | ETF1 | 6770075 | 18177 | -0.816 | -0.4194 | Yes | ||
| 207 | ITGB1 | 5080156 6270528 | 18199 | -0.831 | -0.4044 | Yes | ||
| 208 | UBE2G1 | 2230546 5420528 7040463 | 18223 | -0.851 | -0.3892 | Yes | ||
| 209 | APLP2 | 3610037 | 18272 | -0.911 | -0.3743 | Yes | ||
| 210 | PIAS1 | 5340301 | 18308 | -0.946 | -0.3579 | Yes | ||
| 211 | CLPX | 6220156 | 18313 | -0.958 | -0.3396 | Yes | ||
| 212 | RHOT1 | 6760692 | 18360 | -1.018 | -0.3224 | Yes | ||
| 213 | DAB2IP | 3170064 6200441 | 18365 | -1.031 | -0.3027 | Yes | ||
| 214 | TBK1 | 5690338 | 18393 | -1.069 | -0.2835 | Yes | ||
| 215 | MCL1 | 1660672 | 18416 | -1.131 | -0.2629 | Yes | ||
| 216 | AP3S1 | 5570044 | 18425 | -1.149 | -0.2411 | Yes | ||
| 217 | ATP2A2 | 1090075 3990279 | 18482 | -1.356 | -0.2179 | Yes | ||
| 218 | PPP3CB | 6020156 | 18526 | -1.527 | -0.1908 | Yes | ||
| 219 | HMGCS1 | 6620452 | 18530 | -1.546 | -0.1611 | Yes | ||
| 220 | BTG1 | 4200735 6040131 6200133 | 18602 | -2.662 | -0.1135 | Yes | ||
| 221 | ARID1A | 2630022 1690551 4810110 | 18616 | -5.910 | -0.0000 | Yes |

