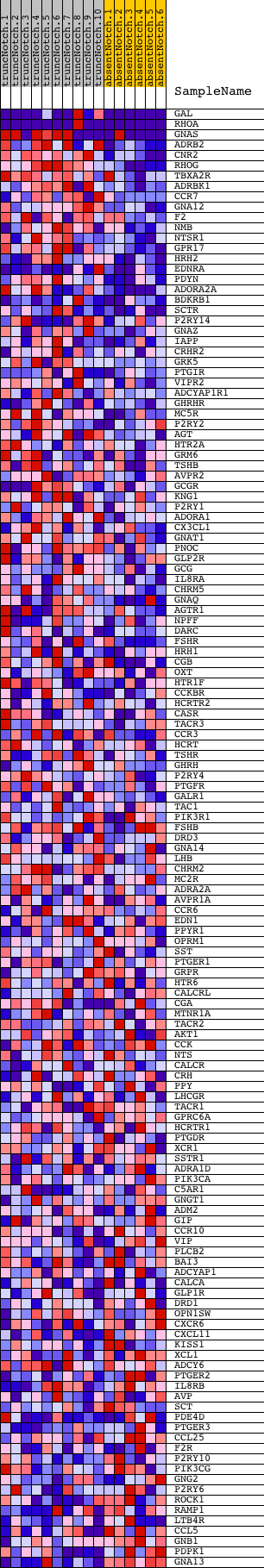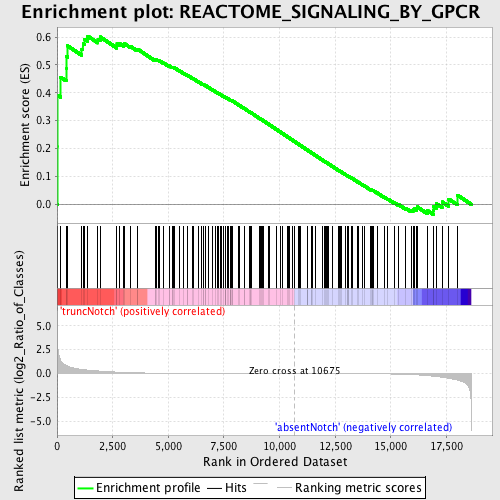
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #truncNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#truncNotch_versus_absentNotch |
| Upregulated in class | truncNotch |
| GeneSet | REACTOME_SIGNALING_BY_GPCR |
| Enrichment Score (ES) | 0.6032162 |
| Normalized Enrichment Score (NES) | 1.505943 |
| Nominal p-value | 0.0018382353 |
| FDR q-value | 0.4223478 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GAL | 3930279 | 15 | 3.926 | 0.2059 | Yes | ||
| 2 | RHOA | 580142 5900131 5340450 | 18 | 3.530 | 0.3917 | Yes | ||
| 3 | GNAS | 630441 1850373 4050152 | 159 | 1.366 | 0.4561 | Yes | ||
| 4 | ADRB2 | 3290373 | 408 | 0.843 | 0.4870 | Yes | ||
| 5 | CNR2 | 2760398 | 415 | 0.835 | 0.5306 | Yes | ||
| 6 | RHOG | 6760575 | 474 | 0.778 | 0.5685 | Yes | ||
| 7 | TBXA2R | 2370292 3940066 | 1108 | 0.429 | 0.5568 | Yes | ||
| 8 | ADRBK1 | 1340333 | 1163 | 0.412 | 0.5756 | Yes | ||
| 9 | CCR7 | 2060687 | 1240 | 0.388 | 0.5919 | Yes | ||
| 10 | GNA12 | 1230301 | 1378 | 0.355 | 0.6032 | Yes | ||
| 11 | F2 | 5720280 | 1835 | 0.270 | 0.5928 | No | ||
| 12 | NMB | 3360735 | 1940 | 0.252 | 0.6004 | No | ||
| 13 | NTSR1 | 6510136 | 2685 | 0.148 | 0.5679 | No | ||
| 14 | GPR17 | 5890671 | 2690 | 0.146 | 0.5754 | No | ||
| 15 | HRH2 | 3840440 | 2783 | 0.135 | 0.5776 | No | ||
| 16 | EDNRA | 6900133 | 2996 | 0.114 | 0.5721 | No | ||
| 17 | PDYN | 5720408 | 3029 | 0.112 | 0.5763 | No | ||
| 18 | ADORA2A | 1990687 | 3284 | 0.090 | 0.5673 | No | ||
| 19 | BDKRB1 | 730592 | 3600 | 0.073 | 0.5540 | No | ||
| 20 | SCTR | 2940070 | 3626 | 0.071 | 0.5564 | No | ||
| 21 | P2RY14 | 6100497 | 4401 | 0.045 | 0.5169 | No | ||
| 22 | GNAZ | 6130296 | 4438 | 0.044 | 0.5172 | No | ||
| 23 | IAPP | 4230040 | 4451 | 0.043 | 0.5189 | No | ||
| 24 | CRHR2 | 4590672 | 4538 | 0.041 | 0.5164 | No | ||
| 25 | GRK5 | 1940348 4670053 | 4616 | 0.039 | 0.5143 | No | ||
| 26 | PTGIR | 2100563 | 4785 | 0.036 | 0.5070 | No | ||
| 27 | VIPR2 | 6520050 | 5033 | 0.031 | 0.4953 | No | ||
| 28 | ADCYAP1R1 | 3390020 4280184 5360180 | 5034 | 0.031 | 0.4970 | No | ||
| 29 | GHRHR | 3130500 4010154 | 5183 | 0.028 | 0.4905 | No | ||
| 30 | MC5R | 2640176 | 5203 | 0.028 | 0.4909 | No | ||
| 31 | P2RY2 | 2640180 2810128 | 5228 | 0.028 | 0.4911 | No | ||
| 32 | AGT | 7000575 | 5282 | 0.027 | 0.4896 | No | ||
| 33 | HTR2A | 360037 6330255 | 5512 | 0.024 | 0.4785 | No | ||
| 34 | GRM6 | 2030398 | 5689 | 0.022 | 0.4701 | No | ||
| 35 | TSHB | 6770500 | 5852 | 0.020 | 0.4624 | No | ||
| 36 | AVPR2 | 360064 | 5859 | 0.020 | 0.4632 | No | ||
| 37 | GCGR | 6620497 | 6092 | 0.018 | 0.4516 | No | ||
| 38 | KNG1 | 6400576 6770347 | 6117 | 0.018 | 0.4512 | No | ||
| 39 | P2RY1 | 6040121 | 6351 | 0.016 | 0.4394 | No | ||
| 40 | ADORA1 | 6370520 | 6511 | 0.015 | 0.4316 | No | ||
| 41 | CX3CL1 | 3990707 | 6572 | 0.014 | 0.4291 | No | ||
| 42 | GNAT1 | 5270048 6100170 | 6581 | 0.014 | 0.4294 | No | ||
| 43 | PNOC | 1990114 | 6654 | 0.014 | 0.4262 | No | ||
| 44 | GLP2R | 4150577 5290273 7050095 | 6690 | 0.013 | 0.4250 | No | ||
| 45 | GCG | 6220735 | 6803 | 0.013 | 0.4197 | No | ||
| 46 | IL8RA | 1580100 | 6970 | 0.012 | 0.4113 | No | ||
| 47 | CHRM5 | 5390333 | 7102 | 0.011 | 0.4048 | No | ||
| 48 | GNAQ | 430670 4210131 5900736 | 7211 | 0.011 | 0.3995 | No | ||
| 49 | AGTR1 | 4780524 2680592 | 7271 | 0.010 | 0.3969 | No | ||
| 50 | NPFF | 2030280 | 7344 | 0.010 | 0.3935 | No | ||
| 51 | DARC | 3940050 | 7383 | 0.010 | 0.3920 | No | ||
| 52 | FSHR | 610041 7040121 | 7473 | 0.009 | 0.3876 | No | ||
| 53 | HRH1 | 840100 | 7556 | 0.009 | 0.3837 | No | ||
| 54 | CGB | 580273 | 7559 | 0.009 | 0.3840 | No | ||
| 55 | OXT | 3850692 | 7569 | 0.009 | 0.3840 | No | ||
| 56 | HTR1F | 670148 | 7643 | 0.009 | 0.3805 | No | ||
| 57 | CCKBR | 2760128 | 7703 | 0.009 | 0.3778 | No | ||
| 58 | HCRTR2 | 2350463 | 7783 | 0.008 | 0.3740 | No | ||
| 59 | CASR | 610504 | 7801 | 0.008 | 0.3735 | No | ||
| 60 | TACR3 | 4780017 | 7805 | 0.008 | 0.3737 | No | ||
| 61 | CCR3 | 50427 | 7836 | 0.008 | 0.3725 | No | ||
| 62 | HCRT | 2970463 | 7861 | 0.008 | 0.3717 | No | ||
| 63 | TSHR | 1190687 5270364 | 7895 | 0.008 | 0.3703 | No | ||
| 64 | GHRH | 4570575 | 8154 | 0.007 | 0.3567 | No | ||
| 65 | P2RY4 | 630053 | 8214 | 0.007 | 0.3539 | No | ||
| 66 | PTGFR | 3850373 | 8424 | 0.006 | 0.3429 | No | ||
| 67 | GALR1 | 6020452 | 8425 | 0.006 | 0.3432 | No | ||
| 68 | TAC1 | 7000195 380706 | 8648 | 0.005 | 0.3315 | No | ||
| 69 | PIK3R1 | 4730671 | 8693 | 0.005 | 0.3294 | No | ||
| 70 | FSHB | 7100592 | 8708 | 0.005 | 0.3289 | No | ||
| 71 | DRD3 | 4780402 | 8723 | 0.005 | 0.3284 | No | ||
| 72 | GNA14 | 3390017 | 9095 | 0.004 | 0.3085 | No | ||
| 73 | LHB | 1450368 | 9149 | 0.004 | 0.3059 | No | ||
| 74 | CHRM2 | 870750 | 9168 | 0.004 | 0.3051 | No | ||
| 75 | MC2R | 2970504 | 9228 | 0.004 | 0.3021 | No | ||
| 76 | ADRA2A | 5340520 | 9282 | 0.004 | 0.2994 | No | ||
| 77 | AVPR1A | 2120300 | 9503 | 0.003 | 0.2877 | No | ||
| 78 | CCR6 | 5720368 6020176 | 9558 | 0.003 | 0.2849 | No | ||
| 79 | EDN1 | 1770047 | 9874 | 0.002 | 0.2679 | No | ||
| 80 | PPYR1 | 1410072 | 10050 | 0.002 | 0.2586 | No | ||
| 81 | OPRM1 | 5360279 | 10143 | 0.001 | 0.2536 | No | ||
| 82 | SST | 6590142 | 10356 | 0.001 | 0.2422 | No | ||
| 83 | PTGER1 | 6100132 | 10411 | 0.001 | 0.2393 | No | ||
| 84 | GRPR | 6020170 | 10462 | 0.000 | 0.2366 | No | ||
| 85 | HTR6 | 4850022 | 10564 | 0.000 | 0.2312 | No | ||
| 86 | CALCRL | 4280035 | 10650 | 0.000 | 0.2266 | No | ||
| 87 | CGA | 630403 | 10846 | -0.000 | 0.2161 | No | ||
| 88 | MTNR1A | 380204 | 10914 | -0.001 | 0.2125 | No | ||
| 89 | TACR2 | 1740358 | 10945 | -0.001 | 0.2109 | No | ||
| 90 | AKT1 | 5290746 | 11232 | -0.001 | 0.1955 | No | ||
| 91 | CCK | 6370368 | 11271 | -0.002 | 0.1935 | No | ||
| 92 | NTS | 380300 2120592 | 11275 | -0.002 | 0.1934 | No | ||
| 93 | CALCR | 1690494 | 11412 | -0.002 | 0.1861 | No | ||
| 94 | CRH | 3710301 | 11433 | -0.002 | 0.1852 | No | ||
| 95 | PPY | 2340373 | 11495 | -0.002 | 0.1820 | No | ||
| 96 | LHCGR | 4560465 5860551 | 11616 | -0.002 | 0.1756 | No | ||
| 97 | TACR1 | 70358 3840411 | 11949 | -0.004 | 0.1578 | No | ||
| 98 | GPRC6A | 4560576 | 12011 | -0.004 | 0.1547 | No | ||
| 99 | HCRTR1 | 1580273 | 12055 | -0.004 | 0.1526 | No | ||
| 100 | PTGDR | 3850161 | 12103 | -0.004 | 0.1503 | No | ||
| 101 | XCR1 | 5080072 | 12160 | -0.004 | 0.1475 | No | ||
| 102 | SSTR1 | 2630471 | 12202 | -0.004 | 0.1455 | No | ||
| 103 | ADRA1D | 380035 540025 | 12382 | -0.005 | 0.1360 | No | ||
| 104 | PIK3CA | 6220129 | 12641 | -0.006 | 0.1224 | No | ||
| 105 | C5AR1 | 4540402 | 12688 | -0.006 | 0.1202 | No | ||
| 106 | GNGT1 | 5220156 | 12750 | -0.006 | 0.1173 | No | ||
| 107 | ADM2 | 4060102 | 12778 | -0.007 | 0.1162 | No | ||
| 108 | GIP | 3170364 | 12976 | -0.008 | 0.1059 | No | ||
| 109 | CCR10 | 4050097 | 13047 | -0.008 | 0.1025 | No | ||
| 110 | VIP | 2850647 | 13054 | -0.008 | 0.1026 | No | ||
| 111 | PLCB2 | 360132 | 13096 | -0.008 | 0.1008 | No | ||
| 112 | BAI3 | 940692 | 13104 | -0.008 | 0.1009 | No | ||
| 113 | ADCYAP1 | 520086 | 13233 | -0.009 | 0.0944 | No | ||
| 114 | CALCA | 5860167 | 13277 | -0.009 | 0.0926 | No | ||
| 115 | GLP1R | 6420528 | 13481 | -0.011 | 0.0822 | No | ||
| 116 | DRD1 | 430025 | 13526 | -0.011 | 0.0803 | No | ||
| 117 | OPN1SW | 6420377 | 13723 | -0.013 | 0.0704 | No | ||
| 118 | CXCR6 | 3190440 | 13812 | -0.014 | 0.0664 | No | ||
| 119 | CXCL11 | 1090551 | 14075 | -0.017 | 0.0531 | No | ||
| 120 | KISS1 | 1580398 | 14111 | -0.018 | 0.0521 | No | ||
| 121 | XCL1 | 3800504 | 14159 | -0.018 | 0.0505 | No | ||
| 122 | ADCY6 | 450364 6290670 6940286 | 14168 | -0.018 | 0.0511 | No | ||
| 123 | PTGER2 | 3170039 | 14216 | -0.019 | 0.0495 | No | ||
| 124 | IL8RB | 450592 1170537 | 14391 | -0.023 | 0.0413 | No | ||
| 125 | AVP | 2100113 | 14708 | -0.032 | 0.0259 | No | ||
| 126 | SCT | 2230348 | 14872 | -0.038 | 0.0190 | No | ||
| 127 | PDE4D | 2470528 6660014 | 15170 | -0.052 | 0.0057 | No | ||
| 128 | PTGER3 | 2900739 6770750 | 15341 | -0.062 | -0.0002 | No | ||
| 129 | CCL25 | 450541 540435 | 15668 | -0.084 | -0.0134 | No | ||
| 130 | F2R | 4810180 | 15920 | -0.107 | -0.0214 | No | ||
| 131 | P2RY10 | 6370039 6840204 | 16000 | -0.117 | -0.0195 | No | ||
| 132 | PIK3CG | 5890110 | 16054 | -0.122 | -0.0160 | No | ||
| 133 | GNG2 | 2230390 | 16167 | -0.135 | -0.0150 | No | ||
| 134 | P2RY6 | 5290400 | 16181 | -0.137 | -0.0085 | No | ||
| 135 | ROCK1 | 130044 | 16649 | -0.218 | -0.0223 | No | ||
| 136 | RAMP1 | 2320168 | 16906 | -0.276 | -0.0216 | No | ||
| 137 | LTB4R | 1770056 | 16926 | -0.282 | -0.0077 | No | ||
| 138 | CCL5 | 3710397 | 17054 | -0.321 | 0.0023 | No | ||
| 139 | GNB1 | 2120397 | 17307 | -0.386 | 0.0090 | No | ||
| 140 | PDPK1 | 6650168 | 17599 | -0.496 | 0.0193 | No | ||
| 141 | GNA13 | 4590102 | 17993 | -0.677 | 0.0337 | No |