Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | absentNotch |
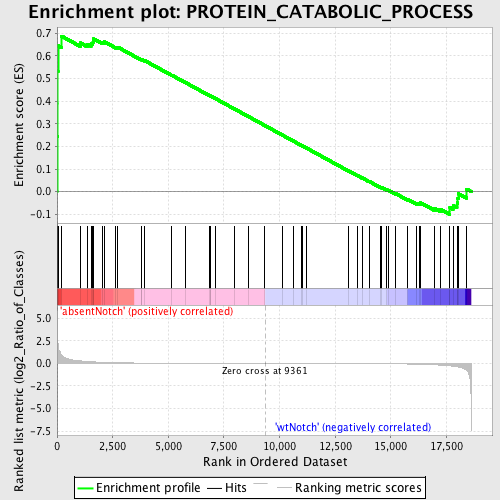
| GeneSet | PROTEIN_CATABOLIC_PROCESS |
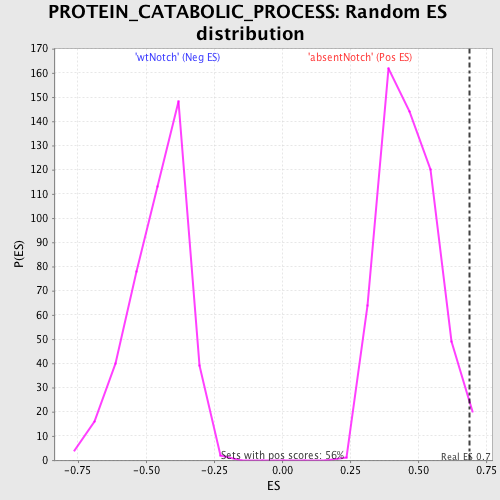
| Enrichment Score (ES) | 0.68743765 |
| Normalized Enrichment Score (NES) | 1.480797 |
| Nominal p-value | 0.019642858 |
| FDR q-value | 0.9437791 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | FAF1 | 2450717 2510121 4730156 | 2 | 4.393 | 0.2443 | Yes | ||
| 2 | UBE2H | 1980142 2970079 | 24 | 2.659 | 0.3910 | Yes | ||
| 3 | EDEM1 | 5270180 5550338 | 25 | 2.560 | 0.5335 | Yes | ||
| 4 | MDM2 | 3450053 5080138 | 44 | 2.014 | 0.6445 | Yes | ||
| 5 | UBE4B | 780008 3610154 | 197 | 0.919 | 0.6874 | Yes | ||
| 6 | RNF11 | 3990068 5080458 | 1038 | 0.275 | 0.6575 | No | ||
| 7 | ANAPC4 | 4540338 | 1353 | 0.214 | 0.6524 | No | ||
| 8 | STUB1 | 5860086 | 1526 | 0.182 | 0.6533 | No | ||
| 9 | UBE2D2 | 4590671 6510446 | 1611 | 0.169 | 0.6582 | No | ||
| 10 | UBE2G1 | 2230546 5420528 7040463 | 1623 | 0.167 | 0.6669 | No | ||
| 11 | PCNP | 610768 | 1642 | 0.165 | 0.6751 | No | ||
| 12 | SMURF2 | 1940161 | 2035 | 0.123 | 0.6608 | No | ||
| 13 | UBE2E1 | 4230064 1090440 | 2122 | 0.116 | 0.6626 | No | ||
| 14 | SMURF1 | 520609 580647 | 2644 | 0.079 | 0.6389 | No | ||
| 15 | SQSTM1 | 6550056 | 2730 | 0.074 | 0.6385 | No | ||
| 16 | UFD1L | 870020 | 3784 | 0.035 | 0.5837 | No | ||
| 17 | FZR1 | 6220504 6660670 | 3800 | 0.035 | 0.5848 | No | ||
| 18 | UBE4A | 6100520 | 3946 | 0.031 | 0.5788 | No | ||
| 19 | CDC23 | 3190593 | 5143 | 0.017 | 0.5153 | No | ||
| 20 | PSMC5 | 2760315 6550021 | 5792 | 0.012 | 0.4811 | No | ||
| 21 | FBXL4 | 3140594 | 6837 | 0.007 | 0.4252 | No | ||
| 22 | FBXO22 | 4760010 | 6887 | 0.007 | 0.4230 | No | ||
| 23 | FRAP1 | 2850037 6660010 | 6904 | 0.007 | 0.4225 | No | ||
| 24 | ARIH1 | 6900025 | 7099 | 0.006 | 0.4124 | No | ||
| 25 | UBE2D1 | 1850400 | 7957 | 0.004 | 0.3665 | No | ||
| 26 | TSG101 | 770148 | 8594 | 0.002 | 0.3323 | No | ||
| 27 | ANAPC5 | 730164 7100685 | 9314 | 0.000 | 0.2936 | No | ||
| 28 | UBE2I | 2680056 6350446 | 10117 | -0.002 | 0.2505 | No | ||
| 29 | UBE2B | 4780497 | 10606 | -0.003 | 0.2244 | No | ||
| 30 | AMFR | 2810041 | 10996 | -0.005 | 0.2037 | No | ||
| 31 | USP33 | 780047 | 11017 | -0.005 | 0.2029 | No | ||
| 32 | PARK2 | 2030053 2030452 3520594 4200136 | 11219 | -0.005 | 0.1923 | No | ||
| 33 | ANAPC10 | 870086 1170037 2260129 | 13109 | -0.014 | 0.0913 | No | ||
| 34 | SIAH2 | 7210672 | 13491 | -0.017 | 0.0718 | No | ||
| 35 | PSMD14 | 5690593 | 13713 | -0.019 | 0.0609 | No | ||
| 36 | VCP | 5340168 | 14027 | -0.023 | 0.0453 | No | ||
| 37 | EGLN2 | 540086 | 14538 | -0.030 | 0.0195 | No | ||
| 38 | MDM4 | 4070504 4780008 | 14602 | -0.031 | 0.0179 | No | ||
| 39 | BTRC | 3170131 | 14812 | -0.035 | 0.0086 | No | ||
| 40 | UBB | 4810138 | 14893 | -0.037 | 0.0064 | No | ||
| 41 | UHRF2 | 60050 840280 1340746 5080091 | 15225 | -0.046 | -0.0089 | No | ||
| 42 | UBE3A | 1240152 2690438 5860609 | 15758 | -0.067 | -0.0338 | No | ||
| 43 | HGS | 5570722 | 16164 | -0.089 | -0.0507 | No | ||
| 44 | UBE2A | 4010563 | 16294 | -0.097 | -0.0523 | No | ||
| 45 | UBE2D3 | 3190452 | 16343 | -0.101 | -0.0493 | No | ||
| 46 | UBE2G2 | 1240035 3870184 | 16958 | -0.153 | -0.0738 | No | ||
| 47 | FBXO7 | 2030397 | 17227 | -0.188 | -0.0778 | No | ||
| 48 | UBE2C | 6130017 | 17653 | -0.271 | -0.0856 | No | ||
| 49 | NEDD8 | 2350722 | 17654 | -0.271 | -0.0706 | No | ||
| 50 | CDC20 | 3440017 3440044 6220088 | 17797 | -0.312 | -0.0609 | No | ||
| 51 | UBE2L3 | 1690707 5900242 | 17975 | -0.375 | -0.0495 | No | ||
| 52 | SYVN1 | 6940594 | 18011 | -0.387 | -0.0299 | No | ||
| 53 | ANAPC2 | 6660438 | 18044 | -0.401 | -0.0093 | No | ||
| 54 | PRSS2 | 1090286 | 18397 | -0.720 | 0.0118 | No |