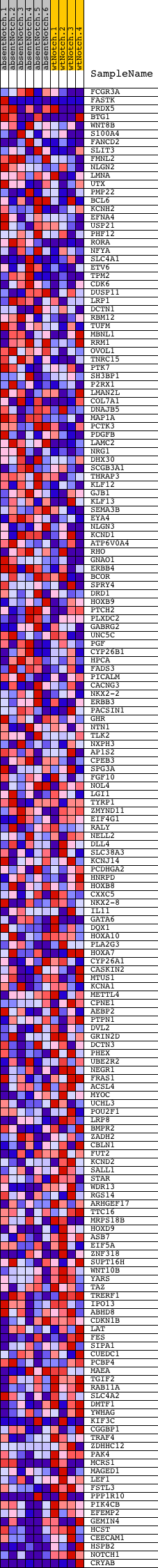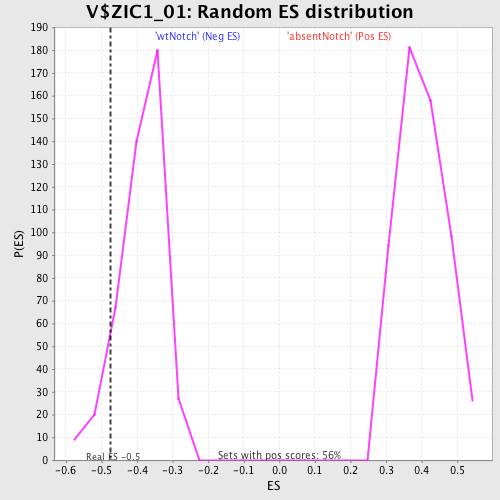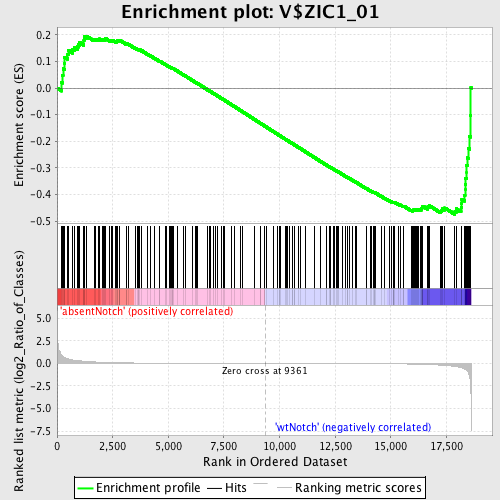
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | V$ZIC1_01 |
| Enrichment Score (ES) | -0.4752585 |
| Normalized Enrichment Score (NES) | -1.225392 |
| Nominal p-value | 0.09255079 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | FCGR3A | 3840184 | 203 | 0.903 | 0.0205 | No | ||
| 2 | FASTK | 2690053 4280288 4610551 6370292 | 232 | 0.847 | 0.0485 | No | ||
| 3 | PRDX5 | 1660592 2030091 | 267 | 0.771 | 0.0735 | No | ||
| 4 | BTG1 | 4200735 6040131 6200133 | 327 | 0.674 | 0.0938 | No | ||
| 5 | WNT8B | 4560301 | 336 | 0.663 | 0.1165 | No | ||
| 6 | S100A4 | 3780113 | 476 | 0.510 | 0.1267 | No | ||
| 7 | FANCD2 | 1850280 6840348 | 518 | 0.486 | 0.1414 | No | ||
| 8 | SLIT3 | 7100132 | 701 | 0.379 | 0.1448 | No | ||
| 9 | FMNL2 | 4060438 | 770 | 0.354 | 0.1535 | No | ||
| 10 | NLGN2 | 5900592 | 925 | 0.305 | 0.1557 | No | ||
| 11 | LMNA | 520471 1500075 3190167 4210020 | 951 | 0.295 | 0.1647 | No | ||
| 12 | UTX | 2900017 6770452 | 1008 | 0.281 | 0.1714 | No | ||
| 13 | PMP22 | 4010239 6550072 | 1203 | 0.240 | 0.1693 | No | ||
| 14 | BCL6 | 940100 | 1204 | 0.240 | 0.1776 | No | ||
| 15 | KCNH2 | 1170451 | 1214 | 0.239 | 0.1855 | No | ||
| 16 | EFNA4 | 4280309 6550576 | 1221 | 0.237 | 0.1934 | No | ||
| 17 | USP21 | 3060446 | 1325 | 0.219 | 0.1955 | No | ||
| 18 | PHF12 | 870400 3130687 | 1663 | 0.162 | 0.1828 | No | ||
| 19 | RORA | 380164 5220215 | 1743 | 0.153 | 0.1839 | No | ||
| 20 | NFYA | 5860368 | 1840 | 0.142 | 0.1836 | No | ||
| 21 | SLC4A1 | 6290577 | 1905 | 0.136 | 0.1849 | No | ||
| 22 | ETV6 | 610524 | 2027 | 0.123 | 0.1826 | No | ||
| 23 | TPM2 | 520735 3870390 | 2100 | 0.117 | 0.1828 | No | ||
| 24 | CDK6 | 4920253 | 2151 | 0.113 | 0.1840 | No | ||
| 25 | DUSP11 | 1500088 4610280 | 2177 | 0.111 | 0.1866 | No | ||
| 26 | LRP1 | 6270386 | 2362 | 0.097 | 0.1800 | No | ||
| 27 | DCTN1 | 1450056 6590022 | 2455 | 0.091 | 0.1782 | No | ||
| 28 | RBM12 | 4570239 6040411 6660129 | 2490 | 0.089 | 0.1794 | No | ||
| 29 | TUFM | 3290685 | 2626 | 0.080 | 0.1749 | No | ||
| 30 | MBNL1 | 2640762 7100048 | 2676 | 0.077 | 0.1749 | No | ||
| 31 | RRM1 | 4150433 | 2692 | 0.076 | 0.1767 | No | ||
| 32 | OVOL1 | 1690131 | 2704 | 0.075 | 0.1787 | No | ||
| 33 | TNRC15 | 6760746 | 2785 | 0.071 | 0.1769 | No | ||
| 34 | PTK7 | 380315 | 2787 | 0.071 | 0.1793 | No | ||
| 35 | SH3BP1 | 3450075 6350288 | 2825 | 0.069 | 0.1797 | No | ||
| 36 | P2RX1 | 2940021 3290338 | 3133 | 0.055 | 0.1649 | No | ||
| 37 | LMAN2L | 50079 6590100 6650338 | 3136 | 0.055 | 0.1668 | No | ||
| 38 | COL7A1 | 1230168 2190072 | 3204 | 0.052 | 0.1649 | No | ||
| 39 | DNAJB5 | 2900215 | 3515 | 0.042 | 0.1496 | No | ||
| 40 | MAP1A | 4920576 | 3622 | 0.039 | 0.1452 | No | ||
| 41 | PCTK3 | 1500328 | 3662 | 0.038 | 0.1444 | No | ||
| 42 | PDGFB | 3060440 6370008 | 3667 | 0.038 | 0.1455 | No | ||
| 43 | LAMC2 | 450692 2680041 4200059 4670148 | 3696 | 0.037 | 0.1452 | No | ||
| 44 | NRG1 | 1050332 | 3813 | 0.035 | 0.1402 | No | ||
| 45 | DHX30 | 2480494 | 4077 | 0.029 | 0.1269 | No | ||
| 46 | SCGB3A1 | 5900333 6400592 | 4184 | 0.028 | 0.1221 | No | ||
| 47 | THRAP3 | 6180309 | 4395 | 0.024 | 0.1116 | No | ||
| 48 | KLF12 | 1660095 4810288 5340546 6520286 | 4618 | 0.022 | 0.1003 | No | ||
| 49 | GJB1 | 1240441 | 4621 | 0.022 | 0.1010 | No | ||
| 50 | KLF13 | 4670563 | 4876 | 0.019 | 0.0878 | No | ||
| 51 | SEMA3B | 5570563 | 4912 | 0.018 | 0.0866 | No | ||
| 52 | EYA4 | 5360286 | 5062 | 0.017 | 0.0791 | No | ||
| 53 | NLGN3 | 2650463 | 5089 | 0.017 | 0.0783 | No | ||
| 54 | KCND1 | 6860278 | 5140 | 0.017 | 0.0762 | No | ||
| 55 | ATP6V0A4 | 360100 | 5177 | 0.016 | 0.0748 | No | ||
| 56 | RHO | 4280398 | 5182 | 0.016 | 0.0751 | No | ||
| 57 | GNAO1 | 2510292 6590487 | 5227 | 0.016 | 0.0733 | No | ||
| 58 | ERBB4 | 4610400 4810017 | 5389 | 0.015 | 0.0651 | No | ||
| 59 | BCOR | 1660452 3940053 | 5685 | 0.013 | 0.0495 | No | ||
| 60 | SPRY4 | 1570594 | 5763 | 0.012 | 0.0458 | No | ||
| 61 | DRD1 | 430025 | 6076 | 0.011 | 0.0292 | No | ||
| 62 | HOXB9 | 2640687 | 6214 | 0.010 | 0.0222 | No | ||
| 63 | PTCH2 | 2900092 | 6274 | 0.010 | 0.0193 | No | ||
| 64 | PLXDC2 | 6400358 | 6307 | 0.010 | 0.0179 | No | ||
| 65 | GABRG2 | 2350402 4210204 6130279 6550037 | 6773 | 0.008 | -0.0070 | No | ||
| 66 | UNC5C | 2640056 | 6844 | 0.007 | -0.0106 | No | ||
| 67 | PGF | 4810593 | 6896 | 0.007 | -0.0131 | No | ||
| 68 | CYP26B1 | 2630142 | 7027 | 0.007 | -0.0199 | No | ||
| 69 | HPCA | 6860010 | 7124 | 0.006 | -0.0249 | No | ||
| 70 | FADS3 | 3120079 | 7227 | 0.006 | -0.0302 | No | ||
| 71 | PICALM | 1940280 | 7377 | 0.005 | -0.0381 | No | ||
| 72 | CACNG3 | 520093 | 7494 | 0.005 | -0.0442 | No | ||
| 73 | NKX2-2 | 4150731 | 7530 | 0.005 | -0.0459 | No | ||
| 74 | ERBB3 | 5890372 | 7825 | 0.004 | -0.0617 | No | ||
| 75 | PACSIN1 | 3870632 6370021 6900707 | 7984 | 0.004 | -0.0702 | No | ||
| 76 | GHR | 2260156 | 8244 | 0.003 | -0.0841 | No | ||
| 77 | NTN1 | 5700600 | 8334 | 0.003 | -0.0889 | No | ||
| 78 | TLK2 | 1170161 | 8891 | 0.001 | -0.1190 | No | ||
| 79 | NXPH3 | 5860292 | 9153 | 0.001 | -0.1331 | No | ||
| 80 | AP1S2 | 780279 4280707 | 9162 | 0.001 | -0.1335 | No | ||
| 81 | CPEB3 | 3940164 | 9319 | 0.000 | -0.1420 | No | ||
| 82 | SPG3A | 2760091 3120170 | 9328 | 0.000 | -0.1424 | No | ||
| 83 | FGF10 | 730458 840706 | 9415 | -0.000 | -0.1471 | No | ||
| 84 | NOL4 | 5050446 5360519 | 9745 | -0.001 | -0.1649 | No | ||
| 85 | LGI1 | 1850020 | 9897 | -0.001 | -0.1730 | No | ||
| 86 | TYRP1 | 3170450 5690204 6290092 | 9997 | -0.002 | -0.1783 | No | ||
| 87 | ZMYND11 | 630181 2570019 4850138 5360040 | 10007 | -0.002 | -0.1788 | No | ||
| 88 | EIF4G1 | 4070446 | 10028 | -0.002 | -0.1798 | No | ||
| 89 | RALY | 940427 1740066 | 10251 | -0.002 | -0.1917 | No | ||
| 90 | NELL2 | 2340725 4280301 5420008 | 10331 | -0.003 | -0.1959 | No | ||
| 91 | DLL4 | 6400403 | 10357 | -0.003 | -0.1972 | No | ||
| 92 | SLC38A3 | 5340142 | 10429 | -0.003 | -0.2010 | No | ||
| 93 | KCNJ14 | 870441 4060113 6110372 | 10573 | -0.003 | -0.2086 | No | ||
| 94 | PCDHGA2 | 1780541 | 10574 | -0.003 | -0.2085 | No | ||
| 95 | HNRPD | 4120021 | 10659 | -0.003 | -0.2129 | No | ||
| 96 | HOXB8 | 3710450 | 10677 | -0.004 | -0.2137 | No | ||
| 97 | CXXC5 | 840010 | 10845 | -0.004 | -0.2226 | No | ||
| 98 | NKX2-8 | 50022 | 10946 | -0.004 | -0.2279 | No | ||
| 99 | IL11 | 1740398 | 11170 | -0.005 | -0.2398 | No | ||
| 100 | GATA6 | 1770010 | 11558 | -0.006 | -0.2606 | No | ||
| 101 | DQX1 | 4150408 | 11848 | -0.007 | -0.2760 | No | ||
| 102 | HOXA10 | 6110397 7100458 | 12089 | -0.008 | -0.2887 | No | ||
| 103 | PLA2G3 | 5130739 | 12100 | -0.008 | -0.2890 | No | ||
| 104 | HOXA7 | 5910152 | 12252 | -0.009 | -0.2968 | No | ||
| 105 | CYP26A1 | 380022 2690600 | 12274 | -0.009 | -0.2977 | No | ||
| 106 | CASKIN2 | 2030113 | 12279 | -0.009 | -0.2975 | No | ||
| 107 | MTUS1 | 780348 4920609 | 12437 | -0.010 | -0.3057 | No | ||
| 108 | KCNA1 | 2030594 | 12467 | -0.010 | -0.3069 | No | ||
| 109 | METTL4 | 1050722 7050296 | 12541 | -0.011 | -0.3105 | No | ||
| 110 | CPNE1 | 3440131 | 12619 | -0.011 | -0.3143 | No | ||
| 111 | AEBP2 | 2970603 | 12661 | -0.011 | -0.3161 | No | ||
| 112 | PTPN1 | 2650056 | 12809 | -0.012 | -0.3237 | No | ||
| 113 | DVL2 | 6110162 | 12968 | -0.013 | -0.3318 | No | ||
| 114 | GRIN2D | 6620372 | 13030 | -0.014 | -0.3346 | No | ||
| 115 | DCTN3 | 6840131 | 13072 | -0.014 | -0.3364 | No | ||
| 116 | PHEX | 540162 | 13134 | -0.014 | -0.3392 | No | ||
| 117 | UBE2R2 | 1230504 | 13262 | -0.015 | -0.3456 | No | ||
| 118 | NEGR1 | 1940731 5130170 | 13432 | -0.016 | -0.3542 | No | ||
| 119 | FRAS1 | 5080170 5670347 | 13457 | -0.017 | -0.3549 | No | ||
| 120 | ACSL4 | 510020 1240041 3190050 4540500 5570463 | 13891 | -0.021 | -0.3776 | No | ||
| 121 | MYOC | 1940270 2100333 | 13893 | -0.021 | -0.3769 | No | ||
| 122 | UCHL3 | 1240735 | 14073 | -0.023 | -0.3858 | No | ||
| 123 | POU2F1 | 70577 430373 4850324 5910056 | 14111 | -0.024 | -0.3870 | No | ||
| 124 | LRP8 | 3610746 5360035 | 14223 | -0.025 | -0.3922 | No | ||
| 125 | BMPR2 | 5340066 6960328 | 14260 | -0.026 | -0.3932 | No | ||
| 126 | ZADH2 | 520438 | 14276 | -0.026 | -0.3931 | No | ||
| 127 | CBLN1 | 5670358 | 14315 | -0.026 | -0.3943 | No | ||
| 128 | FUT2 | 3990047 | 14576 | -0.031 | -0.4073 | No | ||
| 129 | KCND2 | 1170128 2100112 | 14730 | -0.034 | -0.4144 | No | ||
| 130 | SALL1 | 5420020 7050195 | 14926 | -0.038 | -0.4236 | No | ||
| 131 | STAR | 7040670 | 15013 | -0.041 | -0.4269 | No | ||
| 132 | WDR13 | 3840520 4670064 | 15102 | -0.043 | -0.4302 | No | ||
| 133 | RGS14 | 380086 | 15109 | -0.043 | -0.4290 | No | ||
| 134 | ARHGEF17 | 2060008 | 15135 | -0.044 | -0.4288 | No | ||
| 135 | TTC16 | 4830347 | 15172 | -0.045 | -0.4292 | No | ||
| 136 | MRPS18B | 5570086 | 15329 | -0.050 | -0.4359 | No | ||
| 137 | HOXD9 | 2570181 | 15429 | -0.054 | -0.4394 | No | ||
| 138 | ASB7 | 520242 2450088 3990563 4280538 | 15556 | -0.059 | -0.4442 | No | ||
| 139 | EIF5A | 1500059 5290022 6370026 | 15583 | -0.060 | -0.4435 | No | ||
| 140 | ZNF318 | 5890091 6130021 | 15924 | -0.075 | -0.4593 | No | ||
| 141 | SUPT16H | 1240333 | 15987 | -0.078 | -0.4600 | No | ||
| 142 | WNT10B | 510050 | 16005 | -0.079 | -0.4581 | No | ||
| 143 | YARS | 70739 | 16063 | -0.083 | -0.4583 | No | ||
| 144 | TAZ | 7100193 | 16109 | -0.086 | -0.4578 | No | ||
| 145 | TRERF1 | 6370017 6940138 | 16133 | -0.087 | -0.4560 | No | ||
| 146 | IPO13 | 3870088 | 16219 | -0.091 | -0.4574 | No | ||
| 147 | ABHD8 | 6660082 | 16250 | -0.093 | -0.4558 | No | ||
| 148 | CDKN1B | 3800025 6450044 | 16341 | -0.100 | -0.4572 | No | ||
| 149 | LAT | 3170025 | 16367 | -0.102 | -0.4550 | No | ||
| 150 | FES | 3120129 5550600 | 16388 | -0.104 | -0.4524 | No | ||
| 151 | SIPA1 | 5220687 | 16402 | -0.105 | -0.4495 | No | ||
| 152 | CUEDC1 | 1660463 | 16421 | -0.106 | -0.4468 | No | ||
| 153 | PCBP4 | 6350722 | 16439 | -0.108 | -0.4439 | No | ||
| 154 | MAEA | 1170392 | 16646 | -0.123 | -0.4508 | No | ||
| 155 | TGIF2 | 3840364 4050520 | 16656 | -0.123 | -0.4470 | No | ||
| 156 | RAB11A | 5360079 | 16709 | -0.129 | -0.4453 | No | ||
| 157 | SLC4A2 | 2690114 | 16738 | -0.131 | -0.4423 | No | ||
| 158 | DMTF1 | 5910047 1570497 2480100 6620239 | 17235 | -0.189 | -0.4626 | No | ||
| 159 | YWHAG | 3780341 | 17293 | -0.202 | -0.4586 | No | ||
| 160 | KIF3C | 2360162 2940110 | 17332 | -0.209 | -0.4534 | No | ||
| 161 | CGGBP1 | 4810053 | 17430 | -0.226 | -0.4508 | No | ||
| 162 | TRAF4 | 3060041 4920528 6980286 | 17882 | -0.340 | -0.4634 | Yes | ||
| 163 | ZDHHC12 | 3450377 | 17934 | -0.359 | -0.4537 | Yes | ||
| 164 | PAK4 | 1690152 | 18156 | -0.461 | -0.4496 | Yes | ||
| 165 | MCRS1 | 110121 2340592 5270484 | 18175 | -0.474 | -0.4340 | Yes | ||
| 166 | MAGED1 | 6650332 | 18191 | -0.483 | -0.4180 | Yes | ||
| 167 | LEF1 | 2470082 7100288 | 18318 | -0.604 | -0.4038 | Yes | ||
| 168 | FSTL3 | 5420279 | 18336 | -0.626 | -0.3829 | Yes | ||
| 169 | PPP1R10 | 110136 3170725 3610484 | 18342 | -0.635 | -0.3610 | Yes | ||
| 170 | PIK4CB | 510450 1780452 | 18365 | -0.664 | -0.3391 | Yes | ||
| 171 | EFEMP2 | 5550086 | 18401 | -0.727 | -0.3156 | Yes | ||
| 172 | GEMIN4 | 130278 2230075 | 18405 | -0.731 | -0.2903 | Yes | ||
| 173 | HCST | 2350603 | 18450 | -0.870 | -0.2624 | Yes | ||
| 174 | CEECAM1 | 780605 | 18494 | -1.066 | -0.2276 | Yes | ||
| 175 | HSPB2 | 1990110 | 18539 | -1.389 | -0.1815 | Yes | ||
| 176 | NOTCH1 | 3390114 | 18588 | -2.330 | -0.1029 | Yes | ||
| 177 | CRYAB | 4810619 | 18600 | -2.995 | 0.0009 | Yes |