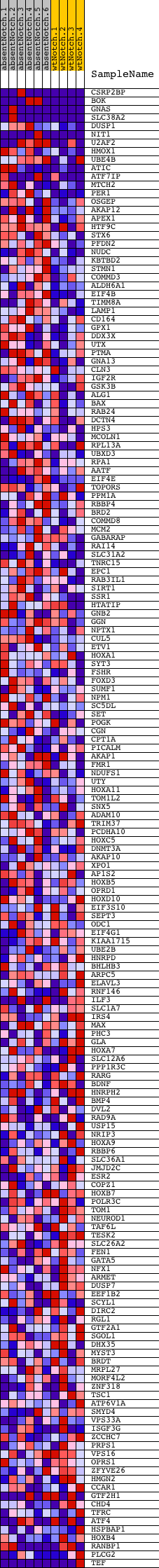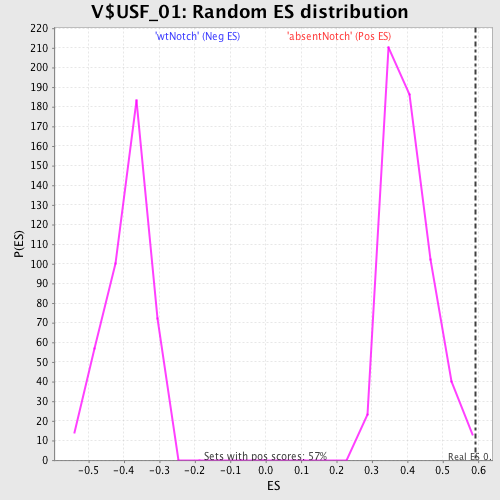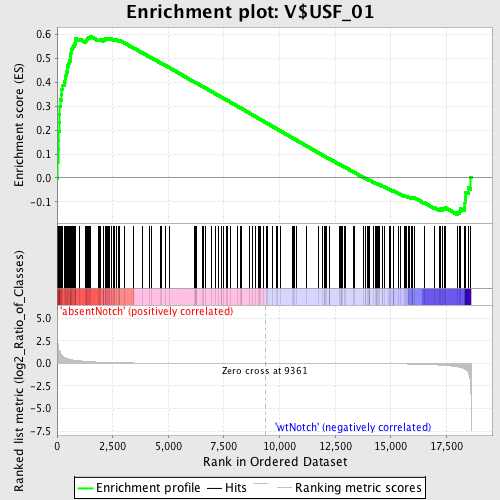
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | V$USF_01 |
| Enrichment Score (ES) | 0.5937556 |
| Normalized Enrichment Score (NES) | 1.480365 |
| Nominal p-value | 0.0034843206 |
| FDR q-value | 0.353288 |
| FWER p-Value | 0.994 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CSRP2BP | 4570717 5550333 | 27 | 2.547 | 0.0644 | Yes | ||
| 2 | BOK | 1170373 5860446 | 40 | 2.181 | 0.1202 | Yes | ||
| 3 | GNAS | 630441 1850373 4050152 | 73 | 1.512 | 0.1576 | Yes | ||
| 4 | SLC38A2 | 1470242 3800026 | 81 | 1.464 | 0.1951 | Yes | ||
| 5 | DUSP1 | 6860121 | 96 | 1.398 | 0.2305 | Yes | ||
| 6 | NIT1 | 1230338 3940671 | 101 | 1.374 | 0.2658 | Yes | ||
| 7 | U2AF2 | 450600 4670128 5420292 | 127 | 1.321 | 0.2987 | Yes | ||
| 8 | HMOX1 | 1740687 | 152 | 1.161 | 0.3274 | Yes | ||
| 9 | UBE4B | 780008 3610154 | 197 | 0.919 | 0.3488 | Yes | ||
| 10 | ATIC | 4010593 6380463 | 213 | 0.887 | 0.3709 | Yes | ||
| 11 | ATF7IP | 2690176 6770021 | 261 | 0.780 | 0.3886 | Yes | ||
| 12 | MTCH2 | 3780603 6620215 | 316 | 0.684 | 0.4033 | Yes | ||
| 13 | PER1 | 3290315 | 355 | 0.638 | 0.4178 | Yes | ||
| 14 | OSGEP | 5700239 | 396 | 0.591 | 0.4309 | Yes | ||
| 15 | AKAP12 | 1450739 | 428 | 0.564 | 0.4438 | Yes | ||
| 16 | APEX1 | 3190519 | 459 | 0.538 | 0.4561 | Yes | ||
| 17 | HTF9C | 3190048 4120132 | 462 | 0.536 | 0.4699 | Yes | ||
| 18 | STX6 | 4850750 6770278 | 527 | 0.480 | 0.4788 | Yes | ||
| 19 | PFDN2 | 4230156 | 538 | 0.473 | 0.4905 | Yes | ||
| 20 | NUDC | 4780025 | 586 | 0.436 | 0.4992 | Yes | ||
| 21 | KBTBD2 | 2320014 | 594 | 0.432 | 0.5100 | Yes | ||
| 22 | STMN1 | 1990717 | 620 | 0.416 | 0.5194 | Yes | ||
| 23 | COMMD3 | 2510259 | 628 | 0.412 | 0.5297 | Yes | ||
| 24 | ALDH6A1 | 1770746 | 655 | 0.400 | 0.5387 | Yes | ||
| 25 | EIF4B | 5390563 5270577 | 692 | 0.385 | 0.5467 | Yes | ||
| 26 | TIMM8A | 110279 | 726 | 0.369 | 0.5544 | Yes | ||
| 27 | LAMP1 | 2470524 | 772 | 0.353 | 0.5611 | Yes | ||
| 28 | CD164 | 3830594 | 817 | 0.336 | 0.5674 | Yes | ||
| 29 | GPX1 | 4150093 | 837 | 0.331 | 0.5750 | Yes | ||
| 30 | DDX3X | 2190020 | 842 | 0.330 | 0.5833 | Yes | ||
| 31 | UTX | 2900017 6770452 | 1008 | 0.281 | 0.5816 | Yes | ||
| 32 | PTMA | 5570148 | 1284 | 0.224 | 0.5725 | Yes | ||
| 33 | GNA13 | 4590102 | 1300 | 0.222 | 0.5774 | Yes | ||
| 34 | CLN3 | 780368 | 1339 | 0.216 | 0.5810 | Yes | ||
| 35 | IGF2R | 1570402 | 1362 | 0.213 | 0.5853 | Yes | ||
| 36 | GSK3B | 5360348 | 1401 | 0.203 | 0.5885 | Yes | ||
| 37 | ALG1 | 2570112 | 1466 | 0.190 | 0.5899 | Yes | ||
| 38 | BAX | 3830008 | 1486 | 0.188 | 0.5938 | Yes | ||
| 39 | RAB24 | 520601 4540739 | 1856 | 0.141 | 0.5774 | No | ||
| 40 | DCTN4 | 3390010 5550170 | 1926 | 0.132 | 0.5771 | No | ||
| 41 | HPS3 | 6650348 | 1933 | 0.131 | 0.5801 | No | ||
| 42 | MCOLN1 | 4480563 | 2065 | 0.120 | 0.5761 | No | ||
| 43 | RPL13A | 2680519 5700142 | 2083 | 0.119 | 0.5783 | No | ||
| 44 | UBXD3 | 5050193 | 2093 | 0.118 | 0.5809 | No | ||
| 45 | RPA1 | 360452 | 2158 | 0.112 | 0.5803 | No | ||
| 46 | AATF | 5220575 | 2175 | 0.111 | 0.5823 | No | ||
| 47 | EIF4E | 1580403 70133 6380215 | 2199 | 0.109 | 0.5839 | No | ||
| 48 | TOPORS | 4060592 | 2257 | 0.105 | 0.5835 | No | ||
| 49 | PPM1A | 1170301 5900273 | 2311 | 0.101 | 0.5833 | No | ||
| 50 | RBBP4 | 6650528 | 2360 | 0.097 | 0.5832 | No | ||
| 51 | BRD2 | 1450300 5340484 6100605 | 2427 | 0.093 | 0.5820 | No | ||
| 52 | COMMD8 | 5890358 | 2553 | 0.084 | 0.5774 | No | ||
| 53 | MCM2 | 5050139 | 2573 | 0.083 | 0.5785 | No | ||
| 54 | GABARAP | 1450286 | 2575 | 0.083 | 0.5806 | No | ||
| 55 | RAI14 | 770288 1660075 | 2653 | 0.078 | 0.5784 | No | ||
| 56 | SLC31A2 | 1450577 | 2778 | 0.071 | 0.5736 | No | ||
| 57 | TNRC15 | 6760746 | 2785 | 0.071 | 0.5751 | No | ||
| 58 | EPC1 | 1570050 5290095 6900193 | 2793 | 0.070 | 0.5765 | No | ||
| 59 | RAB3IL1 | 380685 5420176 | 3017 | 0.060 | 0.5660 | No | ||
| 60 | SIRT1 | 1190731 | 3415 | 0.044 | 0.5456 | No | ||
| 61 | SSR1 | 5890100 | 3855 | 0.033 | 0.5227 | No | ||
| 62 | HTATIP | 5360739 | 4172 | 0.028 | 0.5062 | No | ||
| 63 | GNB2 | 2350053 | 4261 | 0.026 | 0.5021 | No | ||
| 64 | GGN | 1410528 4210082 5290170 | 4660 | 0.021 | 0.4811 | No | ||
| 65 | NPTX1 | 3710736 | 4705 | 0.021 | 0.4793 | No | ||
| 66 | CUL5 | 450142 | 4869 | 0.019 | 0.4709 | No | ||
| 67 | ETV1 | 70735 2940603 5080463 | 5034 | 0.017 | 0.4625 | No | ||
| 68 | HOXA1 | 1190524 5420142 | 6173 | 0.010 | 0.4010 | No | ||
| 69 | SYT3 | 6380017 | 6219 | 0.010 | 0.3988 | No | ||
| 70 | FSHR | 610041 7040121 | 6227 | 0.010 | 0.3987 | No | ||
| 71 | FOXD3 | 6550156 | 6263 | 0.010 | 0.3971 | No | ||
| 72 | SUMF1 | 730050 | 6280 | 0.010 | 0.3965 | No | ||
| 73 | NPM1 | 4730427 | 6525 | 0.009 | 0.3834 | No | ||
| 74 | SC5DL | 6650390 | 6586 | 0.008 | 0.3804 | No | ||
| 75 | SET | 6650286 | 6653 | 0.008 | 0.3770 | No | ||
| 76 | POGK | 5290315 | 6957 | 0.007 | 0.3608 | No | ||
| 77 | CGN | 5890139 6370167 6400537 | 7122 | 0.006 | 0.3521 | No | ||
| 78 | CPT1A | 6350093 | 7263 | 0.006 | 0.3446 | No | ||
| 79 | PICALM | 1940280 | 7377 | 0.005 | 0.3386 | No | ||
| 80 | AKAP1 | 110148 1740735 2260019 7000563 | 7402 | 0.005 | 0.3375 | No | ||
| 81 | FMR1 | 5050075 | 7493 | 0.005 | 0.3327 | No | ||
| 82 | NDUFS1 | 2650253 | 7605 | 0.005 | 0.3268 | No | ||
| 83 | UTY | 5890441 | 7639 | 0.005 | 0.3252 | No | ||
| 84 | HOXA11 | 6980133 | 7789 | 0.004 | 0.3172 | No | ||
| 85 | TOM1L2 | 4810465 | 8126 | 0.003 | 0.2991 | No | ||
| 86 | SNX5 | 1570021 6860609 | 8221 | 0.003 | 0.2940 | No | ||
| 87 | ADAM10 | 3780156 | 8265 | 0.003 | 0.2918 | No | ||
| 88 | TRIM37 | 5270091 | 8308 | 0.003 | 0.2896 | No | ||
| 89 | PCDHA10 | 3830725 | 8631 | 0.002 | 0.2722 | No | ||
| 90 | HOXC5 | 1580017 | 8768 | 0.002 | 0.2648 | No | ||
| 91 | DNMT3A | 4050068 4810717 | 8894 | 0.001 | 0.2581 | No | ||
| 92 | AKAP10 | 2630504 5910577 | 9066 | 0.001 | 0.2488 | No | ||
| 93 | XPO1 | 540707 | 9074 | 0.001 | 0.2485 | No | ||
| 94 | AP1S2 | 780279 4280707 | 9162 | 0.001 | 0.2438 | No | ||
| 95 | HOXB5 | 2900538 | 9273 | 0.000 | 0.2378 | No | ||
| 96 | OPRD1 | 4280138 | 9395 | -0.000 | 0.2312 | No | ||
| 97 | HOXD10 | 6100039 | 9478 | -0.000 | 0.2268 | No | ||
| 98 | EIF3S10 | 1190079 3840176 | 9660 | -0.001 | 0.2170 | No | ||
| 99 | SEPT3 | 3830022 5290079 | 9839 | -0.001 | 0.2074 | No | ||
| 100 | ODC1 | 5670168 | 9886 | -0.001 | 0.2049 | No | ||
| 101 | EIF4G1 | 4070446 | 10028 | -0.002 | 0.1973 | No | ||
| 102 | KIAA1715 | 1190551 | 10592 | -0.003 | 0.1669 | No | ||
| 103 | UBE2B | 4780497 | 10606 | -0.003 | 0.1663 | No | ||
| 104 | HNRPD | 4120021 | 10659 | -0.003 | 0.1635 | No | ||
| 105 | BHLHB3 | 3450438 | 10762 | -0.004 | 0.1581 | No | ||
| 106 | ARPC5 | 1740411 3450300 | 11201 | -0.005 | 0.1345 | No | ||
| 107 | ELAVL3 | 2850014 | 11746 | -0.007 | 0.1052 | No | ||
| 108 | RNF146 | 1660735 | 11931 | -0.008 | 0.0954 | No | ||
| 109 | ILF3 | 940722 3190647 6520110 | 12000 | -0.008 | 0.0919 | No | ||
| 110 | SLC1A7 | 1990181 | 12019 | -0.008 | 0.0911 | No | ||
| 111 | IRS4 | 430341 1190450 | 12085 | -0.008 | 0.0878 | No | ||
| 112 | MAX | 5220021 | 12107 | -0.008 | 0.0869 | No | ||
| 113 | PHC3 | 4920332 | 12231 | -0.009 | 0.0805 | No | ||
| 114 | GLA | 4610364 | 12250 | -0.009 | 0.0797 | No | ||
| 115 | HOXA7 | 5910152 | 12252 | -0.009 | 0.0799 | No | ||
| 116 | SLC12A6 | 610278 1980035 5340671 | 12683 | -0.011 | 0.0569 | No | ||
| 117 | PPP1R3C | 2640170 | 12745 | -0.012 | 0.0539 | No | ||
| 118 | RARG | 6760136 | 12788 | -0.012 | 0.0519 | No | ||
| 119 | BDNF | 2940128 3520368 | 12799 | -0.012 | 0.0517 | No | ||
| 120 | HNRPH2 | 2630040 5720671 | 12824 | -0.012 | 0.0507 | No | ||
| 121 | BMP4 | 380113 | 12935 | -0.013 | 0.0451 | No | ||
| 122 | DVL2 | 6110162 | 12968 | -0.013 | 0.0437 | No | ||
| 123 | RAD9A | 110300 1940632 3990390 6040014 | 13320 | -0.016 | 0.0250 | No | ||
| 124 | USP15 | 610592 3520504 | 13377 | -0.016 | 0.0224 | No | ||
| 125 | NRIP3 | 1340372 | 13782 | -0.020 | 0.0010 | No | ||
| 126 | HOXA9 | 610494 4730040 5130601 | 13846 | -0.021 | -0.0019 | No | ||
| 127 | RBBP6 | 2320129 | 13857 | -0.021 | -0.0019 | No | ||
| 128 | SLC36A1 | 5080242 | 13936 | -0.022 | -0.0055 | No | ||
| 129 | JMJD2C | 3290341 | 13994 | -0.022 | -0.0081 | No | ||
| 130 | ESR2 | 6220138 | 14019 | -0.022 | -0.0088 | No | ||
| 131 | COPZ1 | 6940100 | 14060 | -0.023 | -0.0103 | No | ||
| 132 | HOXB7 | 4540706 4610358 | 14061 | -0.023 | -0.0098 | No | ||
| 133 | POLR3C | 6940546 | 14234 | -0.025 | -0.0184 | No | ||
| 134 | TOM1 | 940398 2190053 | 14302 | -0.026 | -0.0214 | No | ||
| 135 | NEUROD1 | 3060619 | 14357 | -0.027 | -0.0236 | No | ||
| 136 | TAF6L | 1850056 | 14358 | -0.027 | -0.0229 | No | ||
| 137 | TESK2 | 6520524 | 14381 | -0.027 | -0.0234 | No | ||
| 138 | SLC26A2 | 2360278 | 14467 | -0.029 | -0.0272 | No | ||
| 139 | FEN1 | 1770541 | 14511 | -0.030 | -0.0288 | No | ||
| 140 | GATA5 | 430465 | 14616 | -0.032 | -0.0336 | No | ||
| 141 | NFX1 | 450504 | 14641 | -0.032 | -0.0341 | No | ||
| 142 | ARMET | 5130670 | 14699 | -0.033 | -0.0363 | No | ||
| 143 | DUSP7 | 2940309 | 14945 | -0.039 | -0.0486 | No | ||
| 144 | EEF1B2 | 4850400 6130161 | 14998 | -0.040 | -0.0504 | No | ||
| 145 | SCYL1 | 5050717 | 15103 | -0.043 | -0.0549 | No | ||
| 146 | DIRC2 | 2810494 | 15106 | -0.043 | -0.0539 | No | ||
| 147 | RGL1 | 1500097 6900450 | 15139 | -0.044 | -0.0545 | No | ||
| 148 | GTF2A1 | 4120138 4920524 5390110 1230397 | 15351 | -0.051 | -0.0646 | No | ||
| 149 | SGOL1 | 1980075 5220092 6020711 | 15433 | -0.054 | -0.0676 | No | ||
| 150 | DHX35 | 1230440 | 15593 | -0.060 | -0.0747 | No | ||
| 151 | MYST3 | 5270500 | 15679 | -0.063 | -0.0776 | No | ||
| 152 | BRDT | 3940102 | 15695 | -0.064 | -0.0768 | No | ||
| 153 | MRPL27 | 2060131 | 15780 | -0.068 | -0.0796 | No | ||
| 154 | MORF4L2 | 6450133 | 15852 | -0.072 | -0.0816 | No | ||
| 155 | ZNF318 | 5890091 6130021 | 15924 | -0.075 | -0.0835 | No | ||
| 156 | TSC1 | 1850672 | 15956 | -0.077 | -0.0832 | No | ||
| 157 | ATP6V1A | 6590242 | 15966 | -0.077 | -0.0817 | No | ||
| 158 | SMYD4 | 2640403 | 15978 | -0.078 | -0.0803 | No | ||
| 159 | VPS33A | 780113 | 16081 | -0.084 | -0.0836 | No | ||
| 160 | ISGF3G | 3190131 7100142 | 16518 | -0.114 | -0.1043 | No | ||
| 161 | ZCCHC7 | 6510592 | 16946 | -0.151 | -0.1236 | No | ||
| 162 | PRPS1 | 6350129 | 17203 | -0.183 | -0.1327 | No | ||
| 163 | VPS16 | 430064 3390041 6860301 | 17236 | -0.189 | -0.1295 | No | ||
| 164 | OPRS1 | 6130128 6290717 | 17336 | -0.210 | -0.1295 | No | ||
| 165 | ZFYVE26 | 6860152 | 17391 | -0.218 | -0.1268 | No | ||
| 166 | HMGN2 | 3140091 | 17469 | -0.232 | -0.1249 | No | ||
| 167 | CCAR1 | 3190092 | 17984 | -0.379 | -0.1430 | No | ||
| 168 | GTF2H1 | 2570746 2650148 | 18098 | -0.422 | -0.1382 | No | ||
| 169 | CHD4 | 5420059 6130338 6380717 | 18128 | -0.441 | -0.1284 | No | ||
| 170 | TFRC | 4050551 | 18310 | -0.597 | -0.1228 | No | ||
| 171 | ATF4 | 730441 1740195 | 18328 | -0.615 | -0.1078 | No | ||
| 172 | HSPBAP1 | 4060022 | 18335 | -0.623 | -0.0920 | No | ||
| 173 | HOXB4 | 540131 | 18341 | -0.633 | -0.0759 | No | ||
| 174 | RANBP1 | 430215 1090180 | 18348 | -0.644 | -0.0595 | No | ||
| 175 | PLCG2 | 5720008 | 18480 | -0.981 | -0.0413 | No | ||
| 176 | TEF | 1050050 3120397 4060487 | 18575 | -1.877 | 0.0022 | No |