Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | V$SP1_Q2_01 |
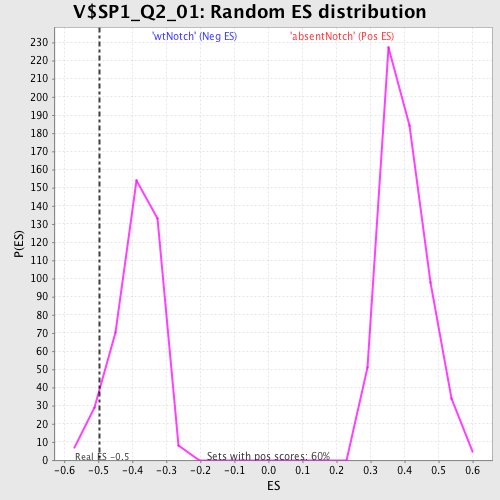
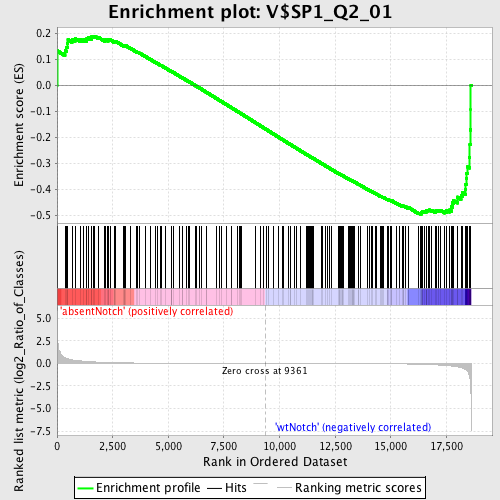
| Enrichment Score (ES) | -0.49670625 |
| Normalized Enrichment Score (NES) | -1.2858177 |
| Nominal p-value | 0.054862842 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | FAF1 | 2450717 2510121 4730156 | 2 | 4.393 | 0.1342 | No | ||
| 2 | PER1 | 3290315 | 355 | 0.638 | 0.1346 | No | ||
| 3 | TPM4 | 2100059 3850072 4920162 | 433 | 0.562 | 0.1477 | No | ||
| 4 | ETV5 | 110017 | 464 | 0.533 | 0.1623 | No | ||
| 5 | TCF12 | 3610324 7000156 | 488 | 0.504 | 0.1765 | No | ||
| 6 | BCL11B | 2680673 | 671 | 0.394 | 0.1787 | No | ||
| 7 | KLF7 | 50390 | 806 | 0.338 | 0.1817 | No | ||
| 8 | ZFP36 | 2030605 | 1033 | 0.277 | 0.1779 | No | ||
| 9 | BCL6 | 940100 | 1204 | 0.240 | 0.1761 | No | ||
| 10 | BMI1 | 3830026 2630204 | 1316 | 0.220 | 0.1768 | No | ||
| 11 | ACLY | 1740609 | 1337 | 0.216 | 0.1823 | No | ||
| 12 | RAP1GDS1 | 50528 | 1396 | 0.204 | 0.1854 | No | ||
| 13 | GRB2 | 6650398 | 1555 | 0.177 | 0.1822 | No | ||
| 14 | SNAG1 | 2760092 | 1557 | 0.177 | 0.1876 | No | ||
| 15 | LASP1 | 1690528 2190341 6420341 | 1635 | 0.166 | 0.1885 | No | ||
| 16 | ANP32A | 510044 1580095 2680037 | 1687 | 0.159 | 0.1906 | No | ||
| 17 | NFE2L1 | 6130053 | 1862 | 0.140 | 0.1854 | No | ||
| 18 | PIP5K2B | 6520156 6620541 | 2139 | 0.114 | 0.1739 | No | ||
| 19 | ARPC4 | 2900056 | 2162 | 0.112 | 0.1762 | No | ||
| 20 | HIPK1 | 110193 | 2248 | 0.105 | 0.1748 | No | ||
| 21 | ARFIP1 | 3130601 | 2307 | 0.101 | 0.1747 | No | ||
| 22 | HS2ST1 | 3170088 | 2309 | 0.101 | 0.1778 | No | ||
| 23 | LDB1 | 5270601 | 2393 | 0.095 | 0.1762 | No | ||
| 24 | HYAL2 | 1500152 | 2560 | 0.084 | 0.1697 | No | ||
| 25 | GABARAP | 1450286 | 2575 | 0.083 | 0.1715 | No | ||
| 26 | THRA | 6770341 110164 1990600 | 2635 | 0.079 | 0.1707 | No | ||
| 27 | SCARF2 | 5290184 | 2993 | 0.061 | 0.1532 | No | ||
| 28 | SH3BP5 | 7100408 | 3022 | 0.060 | 0.1535 | No | ||
| 29 | FUT8 | 1340068 2340056 | 3027 | 0.059 | 0.1551 | No | ||
| 30 | CLPTM1 | 670181 | 3079 | 0.057 | 0.1541 | No | ||
| 31 | KCNN2 | 990025 3520524 | 3312 | 0.048 | 0.1430 | No | ||
| 32 | TBX3 | 2570672 | 3581 | 0.040 | 0.1297 | No | ||
| 33 | CLSTN1 | 6020019 | 3612 | 0.039 | 0.1293 | No | ||
| 34 | BCL7C | 460050 | 3712 | 0.037 | 0.1250 | No | ||
| 35 | KLF16 | 4590725 | 3954 | 0.031 | 0.1129 | No | ||
| 36 | ASCL2 | 1400273 | 4181 | 0.028 | 0.1015 | No | ||
| 37 | PIAS3 | 50204 240739 4060524 | 4411 | 0.024 | 0.0898 | No | ||
| 38 | RNF24 | 4230280 6100070 6620739 | 4519 | 0.023 | 0.0847 | No | ||
| 39 | GGN | 1410528 4210082 5290170 | 4660 | 0.021 | 0.0778 | No | ||
| 40 | CAMK2G | 630097 | 4695 | 0.021 | 0.0765 | No | ||
| 41 | KLF13 | 4670563 | 4876 | 0.019 | 0.0674 | No | ||
| 42 | HSD11B2 | 5900053 | 5120 | 0.017 | 0.0547 | No | ||
| 43 | DUSP15 | 1850044 6960196 | 5151 | 0.017 | 0.0536 | No | ||
| 44 | APLP2 | 3610037 | 5163 | 0.016 | 0.0535 | No | ||
| 45 | GPR3 | 2970452 | 5246 | 0.016 | 0.0495 | No | ||
| 46 | BCL6B | 60047 | 5509 | 0.014 | 0.0357 | No | ||
| 47 | EDG8 | 1990441 | 5633 | 0.013 | 0.0294 | No | ||
| 48 | LOXL4 | 6520095 | 5657 | 0.013 | 0.0286 | No | ||
| 49 | CDH3 | 5390112 6860603 | 5796 | 0.012 | 0.0215 | No | ||
| 50 | TSNAXIP1 | 1770270 6180577 | 5927 | 0.012 | 0.0148 | No | ||
| 51 | PAK1 | 4540315 | 5939 | 0.011 | 0.0145 | No | ||
| 52 | NXPH4 | 3830500 | 6241 | 0.010 | -0.0015 | No | ||
| 53 | PTCH2 | 2900092 | 6274 | 0.010 | -0.0029 | No | ||
| 54 | SP3 | 3840338 | 6417 | 0.009 | -0.0103 | No | ||
| 55 | FGF5 | 5270204 | 6471 | 0.009 | -0.0129 | No | ||
| 56 | KCNB1 | 3610035 | 6712 | 0.008 | -0.0257 | No | ||
| 57 | JUB | 5860711 5900500 | 6732 | 0.008 | -0.0265 | No | ||
| 58 | SOX2 | 630487 2230491 | 7159 | 0.006 | -0.0494 | No | ||
| 59 | ACCN2 | 6450465 | 7311 | 0.006 | -0.0574 | No | ||
| 60 | TIGD4 | 1780114 | 7407 | 0.005 | -0.0624 | No | ||
| 61 | KCNMA1 | 1450402 | 7598 | 0.005 | -0.0726 | No | ||
| 62 | NLK | 2030010 2450041 | 7816 | 0.004 | -0.0842 | No | ||
| 63 | ERBB3 | 5890372 | 7825 | 0.004 | -0.0845 | No | ||
| 64 | ADRB1 | 6860292 | 8096 | 0.003 | -0.0991 | No | ||
| 65 | NUMBL | 6130360 6200333 | 8200 | 0.003 | -0.1046 | No | ||
| 66 | KCNIP2 | 60088 1780324 | 8263 | 0.003 | -0.1078 | No | ||
| 67 | ADAM10 | 3780156 | 8265 | 0.003 | -0.1078 | No | ||
| 68 | DNMT3A | 4050068 4810717 | 8894 | 0.001 | -0.1418 | No | ||
| 69 | NXPH3 | 5860292 | 9153 | 0.001 | -0.1558 | No | ||
| 70 | DES | 1450341 | 9284 | 0.000 | -0.1629 | No | ||
| 71 | GRIN2A | 6550538 | 9404 | -0.000 | -0.1693 | No | ||
| 72 | CACNA1A | 6220164 | 9520 | -0.000 | -0.1755 | No | ||
| 73 | NOL4 | 5050446 5360519 | 9745 | -0.001 | -0.1877 | No | ||
| 74 | CRY1 | 6770039 | 9969 | -0.002 | -0.1997 | No | ||
| 75 | PTHLH | 5290739 | 10151 | -0.002 | -0.2095 | No | ||
| 76 | FEV | 840369 5860139 | 10164 | -0.002 | -0.2100 | No | ||
| 77 | AKT2 | 3850541 3870519 | 10402 | -0.003 | -0.2228 | No | ||
| 78 | PTPN2 | 130315 | 10496 | -0.003 | -0.2278 | No | ||
| 79 | RAB26 | 5910025 | 10510 | -0.003 | -0.2284 | No | ||
| 80 | PTPRJ | 4010707 | 10666 | -0.004 | -0.2367 | No | ||
| 81 | BHLHB3 | 3450438 | 10762 | -0.004 | -0.2417 | No | ||
| 82 | CHAT | 6840603 | 10937 | -0.004 | -0.2510 | No | ||
| 83 | NTF5 | 6110239 | 11188 | -0.005 | -0.2644 | No | ||
| 84 | CLDN11 | 670338 | 11266 | -0.005 | -0.2684 | No | ||
| 85 | FSTL5 | 770280 | 11320 | -0.006 | -0.2711 | No | ||
| 86 | OSBPL9 | 2570373 | 11348 | -0.006 | -0.2724 | No | ||
| 87 | TIMELESS | 3710315 | 11376 | -0.006 | -0.2737 | No | ||
| 88 | GSC | 1580685 | 11421 | -0.006 | -0.2759 | No | ||
| 89 | PHOX2A | 6650121 | 11456 | -0.006 | -0.2776 | No | ||
| 90 | NCOA6 | 1780333 6450110 | 11480 | -0.006 | -0.2787 | No | ||
| 91 | OSBP2 | 6510368 | 11518 | -0.006 | -0.2805 | No | ||
| 92 | MDS1 | 6400072 | 11869 | -0.008 | -0.2992 | No | ||
| 93 | SLC18A3 | 3060035 | 11922 | -0.008 | -0.3018 | No | ||
| 94 | NR2F2 | 3170609 3310577 | 12061 | -0.008 | -0.3090 | No | ||
| 95 | FZD2 | 6480154 | 12135 | -0.009 | -0.3127 | No | ||
| 96 | FGF11 | 840292 | 12253 | -0.009 | -0.3188 | No | ||
| 97 | TBX2 | 1990563 | 12316 | -0.009 | -0.3219 | No | ||
| 98 | POLR2I | 4060400 | 12652 | -0.011 | -0.3397 | No | ||
| 99 | RANBP10 | 2340184 | 12671 | -0.011 | -0.3403 | No | ||
| 100 | ONECUT1 | 6450136 | 12705 | -0.011 | -0.3418 | No | ||
| 101 | TITF1 | 3390554 4540292 | 12732 | -0.012 | -0.3428 | No | ||
| 102 | BDNF | 2940128 3520368 | 12799 | -0.012 | -0.3461 | No | ||
| 103 | SIX4 | 6760022 | 12807 | -0.012 | -0.3461 | No | ||
| 104 | HCN4 | 1050026 | 12830 | -0.012 | -0.3469 | No | ||
| 105 | FGF12 | 1740446 2360037 | 12857 | -0.012 | -0.3479 | No | ||
| 106 | EFNB3 | 5570594 | 13085 | -0.014 | -0.3598 | No | ||
| 107 | CDH2 | 520435 2450451 2760025 6650477 | 13151 | -0.014 | -0.3629 | No | ||
| 108 | ENAH | 1690292 5700300 | 13196 | -0.015 | -0.3648 | No | ||
| 109 | SLC35F5 | 2360369 | 13217 | -0.015 | -0.3655 | No | ||
| 110 | CDH24 | 6980592 | 13255 | -0.015 | -0.3670 | No | ||
| 111 | HRH3 | 6180040 | 13286 | -0.015 | -0.3682 | No | ||
| 112 | DLX3 | 3190497 | 13310 | -0.016 | -0.3689 | No | ||
| 113 | ARHGEF19 | 940102 | 13316 | -0.016 | -0.3687 | No | ||
| 114 | CREB1 | 1500717 2230358 3610600 6550601 | 13388 | -0.016 | -0.3721 | No | ||
| 115 | WNT1 | 4780148 | 13567 | -0.018 | -0.3812 | No | ||
| 116 | SLITRK5 | 6590292 | 13568 | -0.018 | -0.3807 | No | ||
| 117 | TAL1 | 7040239 | 13635 | -0.018 | -0.3837 | No | ||
| 118 | RAB2B | 6200091 | 13950 | -0.022 | -0.4001 | No | ||
| 119 | HOXB7 | 4540706 4610358 | 14061 | -0.023 | -0.4053 | No | ||
| 120 | TRIM28 | 380292 | 14147 | -0.024 | -0.4092 | No | ||
| 121 | HCFC1R1 | 6350500 | 14196 | -0.025 | -0.4110 | No | ||
| 122 | CBLN1 | 5670358 | 14315 | -0.026 | -0.4166 | No | ||
| 123 | EN1 | 5360168 7050025 | 14354 | -0.027 | -0.4179 | No | ||
| 124 | EGLN2 | 540086 | 14538 | -0.030 | -0.4269 | No | ||
| 125 | ERG | 50154 1770739 | 14571 | -0.031 | -0.4276 | No | ||
| 126 | SPAG9 | 580497 2510180 4210685 | 14622 | -0.032 | -0.4294 | No | ||
| 127 | MAP3K11 | 7000039 | 14679 | -0.033 | -0.4314 | No | ||
| 128 | CACNA1D | 1740315 4730731 | 14842 | -0.036 | -0.4391 | No | ||
| 129 | EIF2C1 | 6900551 | 14847 | -0.036 | -0.4382 | No | ||
| 130 | ACE | 1780161 4920315 | 14915 | -0.038 | -0.4407 | No | ||
| 131 | WNT5A | 840685 3120152 | 14967 | -0.039 | -0.4422 | No | ||
| 132 | PPRC1 | 1740441 | 15021 | -0.041 | -0.4439 | No | ||
| 133 | TADA3L | 4210070 5720088 6660333 | 15044 | -0.041 | -0.4438 | No | ||
| 134 | NTRK3 | 4050687 4070167 4610026 6100484 | 15238 | -0.047 | -0.4528 | No | ||
| 135 | PHLDA3 | 510398 | 15393 | -0.053 | -0.4596 | No | ||
| 136 | RELB | 1400048 | 15545 | -0.058 | -0.4660 | No | ||
| 137 | ASB7 | 520242 2450088 3990563 4280538 | 15556 | -0.059 | -0.4647 | No | ||
| 138 | NFAT5 | 2510411 5890195 6550152 | 15573 | -0.059 | -0.4638 | No | ||
| 139 | CHKA | 510324 | 15681 | -0.064 | -0.4677 | No | ||
| 140 | PAX2 | 6040270 7000133 | 15775 | -0.068 | -0.4706 | No | ||
| 141 | MAP4K2 | 4200037 | 15809 | -0.069 | -0.4703 | No | ||
| 142 | ADCK1 | 6620707 | 16261 | -0.094 | -0.4919 | No | ||
| 143 | CORO1C | 4570397 | 16351 | -0.101 | -0.4936 | Yes | ||
| 144 | SESN2 | 2370019 | 16378 | -0.103 | -0.4919 | Yes | ||
| 145 | HAP1 | 1240010 4230079 6980195 | 16381 | -0.103 | -0.4888 | Yes | ||
| 146 | SIPA1 | 5220687 | 16402 | -0.105 | -0.4867 | Yes | ||
| 147 | FBXO3 | 6760673 6860131 | 16437 | -0.108 | -0.4852 | Yes | ||
| 148 | CDK5R1 | 3870161 | 16529 | -0.115 | -0.4867 | Yes | ||
| 149 | GAS7 | 2120358 6450494 | 16586 | -0.119 | -0.4861 | Yes | ||
| 150 | ATP2A2 | 1090075 3990279 | 16605 | -0.120 | -0.4834 | Yes | ||
| 151 | LYL1 | 2900242 | 16606 | -0.120 | -0.4797 | Yes | ||
| 152 | GFRA1 | 3610152 | 16675 | -0.126 | -0.4796 | Yes | ||
| 153 | RELA | 3830075 | 16742 | -0.132 | -0.4791 | Yes | ||
| 154 | IRF2BP1 | 3170019 | 16831 | -0.139 | -0.4797 | Yes | ||
| 155 | SNX2 | 2470068 3360364 6510064 | 16997 | -0.157 | -0.4838 | Yes | ||
| 156 | BAD | 2120148 | 17045 | -0.161 | -0.4814 | Yes | ||
| 157 | GGA1 | 70402 380168 | 17124 | -0.172 | -0.4804 | Yes | ||
| 158 | SMARCD1 | 3060193 3850184 6400369 | 17237 | -0.190 | -0.4807 | Yes | ||
| 159 | CGGBP1 | 4810053 | 17430 | -0.226 | -0.4842 | Yes | ||
| 160 | VAMP2 | 2100100 | 17510 | -0.240 | -0.4811 | Yes | ||
| 161 | RHOG | 6760575 | 17625 | -0.262 | -0.4793 | Yes | ||
| 162 | FBXL19 | 4570039 | 17728 | -0.290 | -0.4759 | Yes | ||
| 163 | OAZ2 | 2810110 | 17731 | -0.291 | -0.4672 | Yes | ||
| 164 | FBXO34 | 4010671 5700647 6450026 | 17764 | -0.301 | -0.4597 | Yes | ||
| 165 | BCL3 | 3990440 | 17777 | -0.305 | -0.4510 | Yes | ||
| 166 | SRR | 1190465 3800494 | 17826 | -0.320 | -0.4438 | Yes | ||
| 167 | MXD4 | 4810113 | 17988 | -0.380 | -0.4409 | Yes | ||
| 168 | CAMKK1 | 6370324 | 17989 | -0.380 | -0.4293 | Yes | ||
| 169 | PAK4 | 1690152 | 18156 | -0.461 | -0.4242 | Yes | ||
| 170 | TMEM24 | 5860039 | 18227 | -0.512 | -0.4124 | Yes | ||
| 171 | UPF2 | 4810471 | 18354 | -0.651 | -0.3993 | Yes | ||
| 172 | PIK4CB | 510450 1780452 | 18365 | -0.664 | -0.3795 | Yes | ||
| 173 | BCL9L | 6940053 | 18389 | -0.706 | -0.3592 | Yes | ||
| 174 | EFEMP2 | 5550086 | 18401 | -0.727 | -0.3376 | Yes | ||
| 175 | DDX6 | 2630025 6770408 | 18456 | -0.888 | -0.3133 | Yes | ||
| 176 | HSPB2 | 1990110 | 18539 | -1.389 | -0.2753 | Yes | ||
| 177 | NOTCH3 | 3060603 | 18554 | -1.615 | -0.2267 | Yes | ||
| 178 | TEF | 1050050 3120397 4060487 | 18575 | -1.877 | -0.1704 | Yes | ||
| 179 | SEP15 | 1780451 2060450 5890451 | 18596 | -2.646 | -0.0906 | Yes | ||
| 180 | CRYAB | 4810619 | 18600 | -2.995 | 0.0009 | Yes |