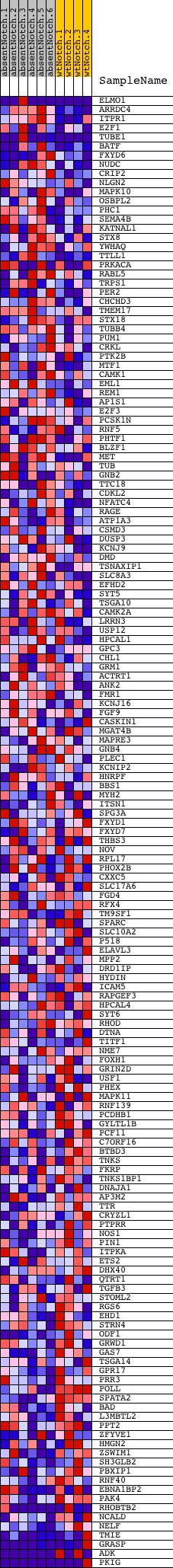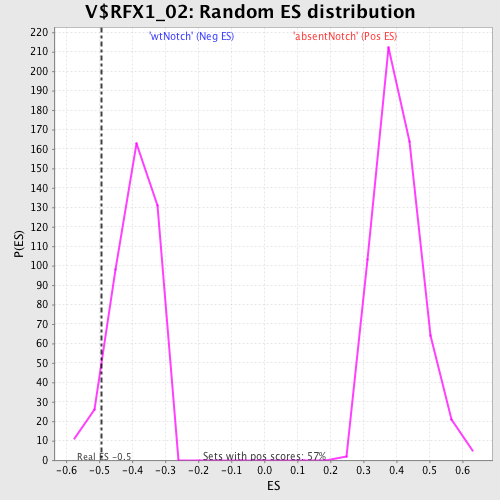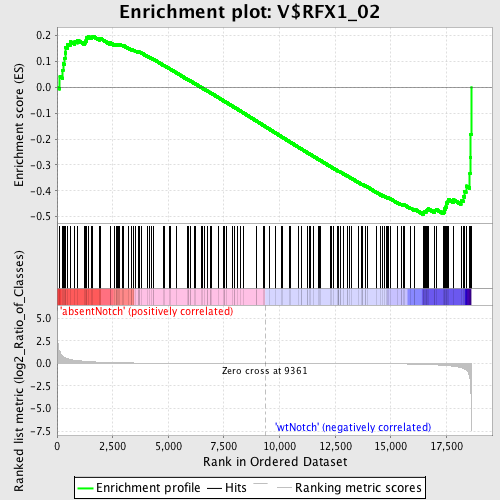
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | V$RFX1_02 |
| Enrichment Score (ES) | -0.4915983 |
| Normalized Enrichment Score (NES) | -1.2462825 |
| Nominal p-value | 0.07692308 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ELMO1 | 110338 1190451 1580148 1850609 2350706 2370369 2450129 3840113 4150184 4280079 6290066 6380435 6770494 7100180 | 128 | 1.319 | 0.0406 | No | ||
| 2 | ARRDC4 | 2810019 | 237 | 0.838 | 0.0650 | No | ||
| 3 | ITPR1 | 3450519 | 269 | 0.768 | 0.0910 | No | ||
| 4 | E2F1 | 5360093 | 341 | 0.657 | 0.1108 | No | ||
| 5 | TUBE1 | 2360079 6520685 | 360 | 0.631 | 0.1326 | No | ||
| 6 | BATF | 6760390 | 376 | 0.615 | 0.1540 | No | ||
| 7 | FXYD6 | 3870164 | 477 | 0.510 | 0.1669 | No | ||
| 8 | NUDC | 4780025 | 586 | 0.436 | 0.1768 | No | ||
| 9 | CRIP2 | 1050750 | 801 | 0.341 | 0.1775 | No | ||
| 10 | NLGN2 | 5900592 | 925 | 0.305 | 0.1818 | No | ||
| 11 | MAPK10 | 6110193 | 1224 | 0.237 | 0.1742 | No | ||
| 12 | OSBPL2 | 5690215 | 1290 | 0.223 | 0.1787 | No | ||
| 13 | PHC1 | 520195 | 1328 | 0.218 | 0.1846 | No | ||
| 14 | SEMA4B | 110347 | 1334 | 0.217 | 0.1922 | No | ||
| 15 | KATNAL1 | 1570538 | 1400 | 0.203 | 0.1960 | No | ||
| 16 | STX8 | 730411 3940427 6290471 | 1563 | 0.176 | 0.1935 | No | ||
| 17 | YWHAQ | 6760524 | 1605 | 0.170 | 0.1974 | No | ||
| 18 | TTLL1 | 6100541 6350079 | 1891 | 0.137 | 0.1870 | No | ||
| 19 | PRKACA | 2640731 4050048 | 1953 | 0.129 | 0.1883 | No | ||
| 20 | RABL5 | 770368 | 2381 | 0.096 | 0.1686 | No | ||
| 21 | TRPS1 | 870100 | 2389 | 0.095 | 0.1717 | No | ||
| 22 | PER2 | 3120204 3390593 | 2588 | 0.082 | 0.1639 | No | ||
| 23 | CHCHD3 | 4200044 | 2658 | 0.078 | 0.1630 | No | ||
| 24 | TMEM17 | 5900079 | 2660 | 0.077 | 0.1657 | No | ||
| 25 | STX18 | 4760706 | 2735 | 0.074 | 0.1644 | No | ||
| 26 | TUBB4 | 5310681 | 2739 | 0.074 | 0.1668 | No | ||
| 27 | PUM1 | 6130500 | 2794 | 0.070 | 0.1665 | No | ||
| 28 | CRKL | 4050427 | 2922 | 0.064 | 0.1619 | No | ||
| 29 | PTK2B | 4730411 | 2982 | 0.062 | 0.1609 | No | ||
| 30 | MTF1 | 520427 2340364 | 3201 | 0.052 | 0.1510 | No | ||
| 31 | CAMK1 | 3390427 5220086 6940504 | 3342 | 0.047 | 0.1451 | No | ||
| 32 | EML1 | 6980332 | 3360 | 0.046 | 0.1458 | No | ||
| 33 | REM1 | 2340112 | 3428 | 0.044 | 0.1438 | No | ||
| 34 | AP1S1 | 2030471 4570520 6900603 | 3532 | 0.041 | 0.1397 | No | ||
| 35 | E2F3 | 50162 460180 | 3661 | 0.038 | 0.1341 | No | ||
| 36 | PCSK1N | 4010435 | 3668 | 0.038 | 0.1351 | No | ||
| 37 | RNF5 | 2760088 | 3689 | 0.037 | 0.1354 | No | ||
| 38 | PHTF1 | 7050441 | 3697 | 0.037 | 0.1363 | No | ||
| 39 | BLZF1 | 630088 | 3796 | 0.035 | 0.1323 | No | ||
| 40 | MET | 2690348 | 4071 | 0.030 | 0.1185 | No | ||
| 41 | TUB | 3990131 6590129 | 4141 | 0.028 | 0.1158 | No | ||
| 42 | GNB2 | 2350053 | 4261 | 0.026 | 0.1103 | No | ||
| 43 | TTC18 | 2450369 3360053 | 4320 | 0.026 | 0.1081 | No | ||
| 44 | CDKL2 | 2230673 2320594 3440048 4230670 | 4330 | 0.025 | 0.1085 | No | ||
| 45 | NFATC4 | 2470735 | 4783 | 0.020 | 0.0847 | No | ||
| 46 | RAGE | 730102 2120706 7050315 | 4804 | 0.019 | 0.0843 | No | ||
| 47 | ATP1A3 | 5690674 | 5061 | 0.017 | 0.0711 | No | ||
| 48 | CSMD3 | 1660427 1940687 | 5105 | 0.017 | 0.0694 | No | ||
| 49 | DUSP3 | 1240129 6840215 | 5357 | 0.015 | 0.0563 | No | ||
| 50 | KCNJ9 | 3440292 | 5868 | 0.012 | 0.0291 | No | ||
| 51 | DMD | 1740041 3990332 | 5912 | 0.012 | 0.0272 | No | ||
| 52 | TSNAXIP1 | 1770270 6180577 | 5927 | 0.012 | 0.0268 | No | ||
| 53 | SLC8A3 | 4120711 6290592 | 5981 | 0.011 | 0.0244 | No | ||
| 54 | EFHD2 | 5050427 | 6011 | 0.011 | 0.0232 | No | ||
| 55 | SYT5 | 4150082 | 6163 | 0.010 | 0.0154 | No | ||
| 56 | TSGA10 | 450594 3130435 4920093 | 6200 | 0.010 | 0.0138 | No | ||
| 57 | CAMK2A | 1740333 1940112 4150292 | 6487 | 0.009 | -0.0014 | No | ||
| 58 | LRRN3 | 1450133 | 6534 | 0.009 | -0.0036 | No | ||
| 59 | USP12 | 6130735 6620280 | 6608 | 0.008 | -0.0072 | No | ||
| 60 | HPCAL1 | 1980129 2350348 | 6758 | 0.008 | -0.0150 | No | ||
| 61 | GPC3 | 6180735 | 6767 | 0.008 | -0.0152 | No | ||
| 62 | CHL1 | 50072 1500021 1580286 6660524 | 6890 | 0.007 | -0.0215 | No | ||
| 63 | GRM1 | 5890324 | 6901 | 0.007 | -0.0218 | No | ||
| 64 | ACTRT1 | 4480347 | 6947 | 0.007 | -0.0240 | No | ||
| 65 | ANK2 | 6510546 | 7245 | 0.006 | -0.0399 | No | ||
| 66 | FMR1 | 5050075 | 7493 | 0.005 | -0.0531 | No | ||
| 67 | KCNJ16 | 4780273 | 7527 | 0.005 | -0.0547 | No | ||
| 68 | FGF9 | 1050195 | 7535 | 0.005 | -0.0549 | No | ||
| 69 | CASKIN1 | 580035 3710500 | 7622 | 0.005 | -0.0594 | No | ||
| 70 | MGAT4B | 130053 6840100 | 7870 | 0.004 | -0.0726 | No | ||
| 71 | MAPRE3 | 7050504 | 7981 | 0.004 | -0.0784 | No | ||
| 72 | GNB4 | 3390079 4230035 | 7988 | 0.004 | -0.0786 | No | ||
| 73 | PLEC1 | 6520292 50500 380075 1570088 2510692 2630497 2680017 2900746 4560497 4850376 6290398 7040471 | 8100 | 0.003 | -0.0845 | No | ||
| 74 | KCNIP2 | 60088 1780324 | 8263 | 0.003 | -0.0932 | No | ||
| 75 | HNRPF | 670138 | 8374 | 0.003 | -0.0991 | No | ||
| 76 | BBS1 | 4480427 | 8971 | 0.001 | -0.1313 | No | ||
| 77 | MYH2 | 2340433 | 8978 | 0.001 | -0.1316 | No | ||
| 78 | ITSN1 | 2640577 | 9297 | 0.000 | -0.1488 | No | ||
| 79 | SPG3A | 2760091 3120170 | 9328 | 0.000 | -0.1505 | No | ||
| 80 | FXYD1 | 1400239 6560129 | 9542 | -0.000 | -0.1620 | No | ||
| 81 | FXYD7 | 3120332 | 9833 | -0.001 | -0.1777 | No | ||
| 82 | THBS3 | 2360465 4050176 | 10081 | -0.002 | -0.1910 | No | ||
| 83 | NOV | 2850746 | 10112 | -0.002 | -0.1925 | No | ||
| 84 | RPL17 | 2690167 5690504 | 10433 | -0.003 | -0.2098 | No | ||
| 85 | PHOX2B | 5270075 | 10500 | -0.003 | -0.2132 | No | ||
| 86 | CXXC5 | 840010 | 10845 | -0.004 | -0.2317 | No | ||
| 87 | SLC17A6 | 4210576 | 10970 | -0.004 | -0.2383 | No | ||
| 88 | FGD4 | 520168 870411 2640253 6550338 6650364 | 10985 | -0.005 | -0.2389 | No | ||
| 89 | RFX4 | 3710100 5340215 | 11260 | -0.005 | -0.2535 | No | ||
| 90 | TM9SF1 | 2760333 | 11276 | -0.005 | -0.2541 | No | ||
| 91 | SPARC | 1690086 | 11335 | -0.006 | -0.2571 | No | ||
| 92 | SLC10A2 | 110537 | 11405 | -0.006 | -0.2606 | No | ||
| 93 | P518 | 3610435 | 11521 | -0.006 | -0.2666 | No | ||
| 94 | ELAVL3 | 2850014 | 11746 | -0.007 | -0.2785 | No | ||
| 95 | MPP2 | 4200438 | 11772 | -0.007 | -0.2796 | No | ||
| 96 | DRD1IP | 1570100 | 11843 | -0.007 | -0.2831 | No | ||
| 97 | HYDIN | 3830427 | 12285 | -0.009 | -0.3067 | No | ||
| 98 | ICAM5 | 2350647 | 12342 | -0.010 | -0.3094 | No | ||
| 99 | RAPGEF3 | 5050592 | 12443 | -0.010 | -0.3144 | No | ||
| 100 | HPCAL4 | 2600484 | 12587 | -0.011 | -0.3218 | No | ||
| 101 | SYT6 | 2510280 3850128 4540064 | 12636 | -0.011 | -0.3240 | No | ||
| 102 | RHOD | 1340128 | 12650 | -0.011 | -0.3243 | No | ||
| 103 | DTNA | 1340600 1780731 2340278 2850132 | 12654 | -0.011 | -0.3241 | No | ||
| 104 | TITF1 | 3390554 4540292 | 12732 | -0.012 | -0.3278 | No | ||
| 105 | NME7 | 60170 1230551 2850333 4200102 4560411 5090647 | 12751 | -0.012 | -0.3284 | No | ||
| 106 | FOXH1 | 2060180 | 12886 | -0.012 | -0.3352 | No | ||
| 107 | GRIN2D | 6620372 | 13030 | -0.014 | -0.3424 | No | ||
| 108 | USF1 | 4280156 4610114 6370113 | 13074 | -0.014 | -0.3443 | No | ||
| 109 | PHEX | 540162 | 13134 | -0.014 | -0.3470 | No | ||
| 110 | MAPK11 | 130452 3120440 3610465 | 13252 | -0.015 | -0.3528 | No | ||
| 111 | RNF139 | 2650114 3610450 | 13550 | -0.018 | -0.3682 | No | ||
| 112 | PCDHB1 | 2260706 | 13670 | -0.019 | -0.3740 | No | ||
| 113 | GYLTL1B | 2120167 6110575 | 13719 | -0.019 | -0.3759 | No | ||
| 114 | PCF11 | 1240180 | 13721 | -0.019 | -0.3753 | No | ||
| 115 | C7ORF16 | 4780215 | 13853 | -0.021 | -0.3816 | No | ||
| 116 | BTBD3 | 1980041 2510577 3190053 4200746 | 13856 | -0.021 | -0.3810 | No | ||
| 117 | TNKS | 2760563 | 13961 | -0.022 | -0.3858 | No | ||
| 118 | FKRP | 130546 | 14372 | -0.027 | -0.4070 | No | ||
| 119 | TNKS1BP1 | 7040184 | 14556 | -0.031 | -0.4159 | No | ||
| 120 | DNAJA1 | 1660010 2320358 6840136 | 14647 | -0.032 | -0.4196 | No | ||
| 121 | AP3M2 | 6370474 | 14718 | -0.034 | -0.4221 | No | ||
| 122 | TTR | 4150072 | 14815 | -0.036 | -0.4261 | No | ||
| 123 | CRYZL1 | 730022 2320044 | 14872 | -0.037 | -0.4278 | No | ||
| 124 | PTPRR | 130121 3990273 | 14884 | -0.037 | -0.4270 | No | ||
| 125 | NOS1 | 5860129 | 14970 | -0.039 | -0.4302 | No | ||
| 126 | PIN1 | 2970408 | 15294 | -0.049 | -0.4460 | No | ||
| 127 | ITPKA | 6220075 | 15457 | -0.055 | -0.4527 | No | ||
| 128 | ETS2 | 360451 | 15465 | -0.055 | -0.4511 | No | ||
| 129 | DHX40 | 360672 | 15558 | -0.059 | -0.4540 | No | ||
| 130 | QTRT1 | 1340162 | 15609 | -0.061 | -0.4545 | No | ||
| 131 | TGFB3 | 1070041 | 15898 | -0.074 | -0.4675 | No | ||
| 132 | STOML2 | 1940487 | 16052 | -0.082 | -0.4728 | No | ||
| 133 | RGS6 | 6040601 | 16076 | -0.084 | -0.4710 | No | ||
| 134 | EHD1 | 3840731 | 16457 | -0.109 | -0.4877 | Yes | ||
| 135 | STRN4 | 3440711 | 16473 | -0.111 | -0.4845 | Yes | ||
| 136 | ODF1 | 6650110 | 16494 | -0.113 | -0.4815 | Yes | ||
| 137 | GRWD1 | 6350528 | 16540 | -0.115 | -0.4798 | Yes | ||
| 138 | GAS7 | 2120358 6450494 | 16586 | -0.119 | -0.4779 | Yes | ||
| 139 | TSGA14 | 6660465 | 16619 | -0.121 | -0.4753 | Yes | ||
| 140 | GPR17 | 5890671 | 16662 | -0.124 | -0.4731 | Yes | ||
| 141 | PRR3 | 3440112 | 16688 | -0.127 | -0.4699 | Yes | ||
| 142 | POLL | 5910673 6350672 | 16961 | -0.153 | -0.4791 | Yes | ||
| 143 | SPATA2 | 1230215 5910541 | 16977 | -0.155 | -0.4744 | Yes | ||
| 144 | BAD | 2120148 | 17045 | -0.161 | -0.4722 | Yes | ||
| 145 | L3MBTL2 | 6020341 | 17367 | -0.215 | -0.4818 | Yes | ||
| 146 | PPT2 | 2340059 3450148 | 17423 | -0.225 | -0.4767 | Yes | ||
| 147 | ZFYVE1 | 60440 | 17427 | -0.225 | -0.4687 | Yes | ||
| 148 | HMGN2 | 3140091 | 17469 | -0.232 | -0.4626 | Yes | ||
| 149 | ZSWIM1 | 7050239 | 17497 | -0.238 | -0.4555 | Yes | ||
| 150 | SH3GLB2 | 2030086 | 17498 | -0.238 | -0.4469 | Yes | ||
| 151 | PBXIP1 | 70131 | 17566 | -0.250 | -0.4416 | Yes | ||
| 152 | RNF40 | 3610397 | 17595 | -0.257 | -0.4338 | Yes | ||
| 153 | EBNA1BP2 | 50369 1400433 6040019 | 17808 | -0.315 | -0.4339 | Yes | ||
| 154 | PAK4 | 1690152 | 18156 | -0.461 | -0.4361 | Yes | ||
| 155 | RHOBTB2 | 1570129 2320253 3520253 | 18270 | -0.551 | -0.4224 | Yes | ||
| 156 | NCALD | 730176 | 18313 | -0.599 | -0.4030 | Yes | ||
| 157 | NELF | 5910026 | 18382 | -0.700 | -0.3815 | Yes | ||
| 158 | TMIE | 2370164 | 18552 | -1.580 | -0.3337 | Yes | ||
| 159 | GRASP | 3940450 5910735 | 18567 | -1.761 | -0.2710 | Yes | ||
| 160 | ADK | 380338 520180 5270524 | 18592 | -2.536 | -0.1808 | Yes | ||
| 161 | PKIG | 2030300 5260255 | 18615 | -5.051 | 0.0001 | Yes |