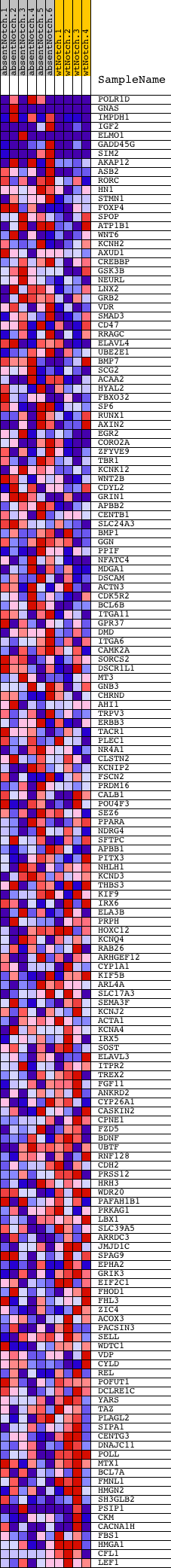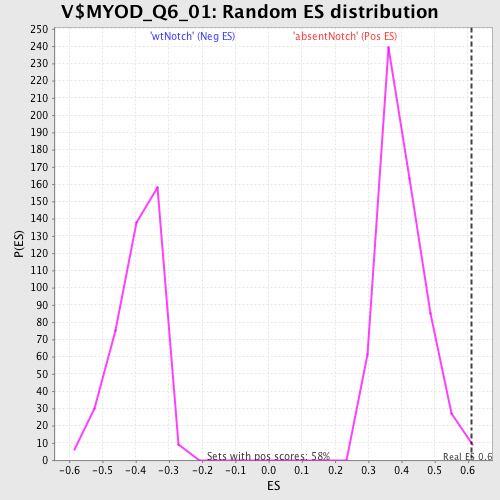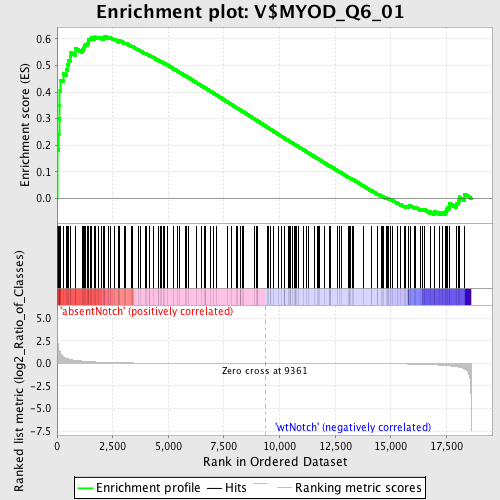
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | V$MYOD_Q6_01 |
| Enrichment Score (ES) | 0.61049575 |
| Normalized Enrichment Score (NES) | 1.5195686 |
| Nominal p-value | 0.006849315 |
| FDR q-value | 0.2870556 |
| FWER p-Value | 0.953 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | POLR1D | 70082 450750 6040458 | 1 | 4.594 | 0.1853 | Yes | ||
| 2 | GNAS | 630441 1850373 4050152 | 73 | 1.512 | 0.2425 | Yes | ||
| 3 | IMPDH1 | 3190735 7050546 | 87 | 1.425 | 0.2992 | Yes | ||
| 4 | IGF2 | 6510020 | 122 | 1.330 | 0.3511 | Yes | ||
| 5 | ELMO1 | 110338 1190451 1580148 1850609 2350706 2370369 2450129 3840113 4150184 4280079 6290066 6380435 6770494 7100180 | 128 | 1.319 | 0.4040 | Yes | ||
| 6 | GADD45G | 2510142 | 166 | 1.064 | 0.4449 | Yes | ||
| 7 | SIM2 | 2100044 | 275 | 0.756 | 0.4696 | Yes | ||
| 8 | AKAP12 | 1450739 | 428 | 0.564 | 0.4841 | Yes | ||
| 9 | ASB2 | 4760168 | 473 | 0.515 | 0.5025 | Yes | ||
| 10 | RORC | 1740121 | 523 | 0.483 | 0.5194 | Yes | ||
| 11 | HN1 | 3360156 | 582 | 0.437 | 0.5339 | Yes | ||
| 12 | STMN1 | 1990717 | 620 | 0.416 | 0.5486 | Yes | ||
| 13 | FOXP4 | 6290022 | 807 | 0.338 | 0.5522 | Yes | ||
| 14 | SPOP | 450035 | 810 | 0.337 | 0.5657 | Yes | ||
| 15 | ATP1B1 | 3130594 | 1123 | 0.256 | 0.5591 | Yes | ||
| 16 | WNT6 | 4570364 | 1181 | 0.244 | 0.5659 | Yes | ||
| 17 | KCNH2 | 1170451 | 1214 | 0.239 | 0.5738 | Yes | ||
| 18 | AXUD1 | 1230053 | 1260 | 0.228 | 0.5805 | Yes | ||
| 19 | CREBBP | 5690035 7040050 | 1349 | 0.214 | 0.5844 | Yes | ||
| 20 | GSK3B | 5360348 | 1401 | 0.203 | 0.5898 | Yes | ||
| 21 | NEURL | 5390538 | 1423 | 0.200 | 0.5968 | Yes | ||
| 22 | LNX2 | 3130025 | 1480 | 0.189 | 0.6014 | Yes | ||
| 23 | GRB2 | 6650398 | 1555 | 0.177 | 0.6045 | Yes | ||
| 24 | VDR | 130156 510438 | 1682 | 0.159 | 0.6041 | Yes | ||
| 25 | SMAD3 | 6450671 | 1739 | 0.154 | 0.6073 | Yes | ||
| 26 | CD47 | 1850603 | 1857 | 0.141 | 0.6066 | Yes | ||
| 27 | RRAGC | 5910348 | 1989 | 0.126 | 0.6046 | Yes | ||
| 28 | ELAVL4 | 50735 3360086 5220167 | 2105 | 0.117 | 0.6031 | Yes | ||
| 29 | UBE2E1 | 4230064 1090440 | 2122 | 0.116 | 0.6069 | Yes | ||
| 30 | BMP7 | 870039 | 2141 | 0.114 | 0.6105 | Yes | ||
| 31 | SCG2 | 5670438 130671 | 2316 | 0.101 | 0.6051 | No | ||
| 32 | ACAA2 | 1570347 2360324 6130139 | 2386 | 0.095 | 0.6052 | No | ||
| 33 | HYAL2 | 1500152 | 2560 | 0.084 | 0.5992 | No | ||
| 34 | FBXO32 | 110037 610750 | 2771 | 0.072 | 0.5908 | No | ||
| 35 | SP6 | 60484 510452 2690333 | 2780 | 0.071 | 0.5932 | No | ||
| 36 | RUNX1 | 3840711 | 2788 | 0.071 | 0.5957 | No | ||
| 37 | AXIN2 | 3850131 | 3041 | 0.059 | 0.5844 | No | ||
| 38 | EGR2 | 3800403 | 3085 | 0.057 | 0.5843 | No | ||
| 39 | CORO2A | 3840605 6620390 | 3329 | 0.047 | 0.5731 | No | ||
| 40 | ZFYVE9 | 6980288 7100017 | 3385 | 0.045 | 0.5719 | No | ||
| 41 | TBR1 | 5130053 | 3645 | 0.038 | 0.5594 | No | ||
| 42 | KCNK12 | 4070072 | 3768 | 0.036 | 0.5542 | No | ||
| 43 | WNT2B | 3130088 5960575 | 3960 | 0.031 | 0.5452 | No | ||
| 44 | CDYL2 | 3450711 | 3995 | 0.031 | 0.5446 | No | ||
| 45 | GRIN1 | 3800014 7000609 | 4163 | 0.028 | 0.5366 | No | ||
| 46 | APBB2 | 5130403 | 4167 | 0.028 | 0.5376 | No | ||
| 47 | CENTB1 | 2120093 | 4324 | 0.025 | 0.5302 | No | ||
| 48 | SLC24A3 | 5080707 | 4537 | 0.023 | 0.5196 | No | ||
| 49 | BMP1 | 380594 2940576 3710593 | 4556 | 0.022 | 0.5195 | No | ||
| 50 | GGN | 1410528 4210082 5290170 | 4660 | 0.021 | 0.5148 | No | ||
| 51 | PPIF | 1740059 | 4674 | 0.021 | 0.5149 | No | ||
| 52 | NFATC4 | 2470735 | 4783 | 0.020 | 0.5099 | No | ||
| 53 | MDGA1 | 1170139 | 4825 | 0.019 | 0.5084 | No | ||
| 54 | DSCAM | 1780050 2450731 2810438 | 4940 | 0.018 | 0.5030 | No | ||
| 55 | ACTN3 | 3140541 6480598 | 5251 | 0.016 | 0.4868 | No | ||
| 56 | CDK5R2 | 3800110 | 5409 | 0.015 | 0.4789 | No | ||
| 57 | BCL6B | 60047 | 5509 | 0.014 | 0.4741 | No | ||
| 58 | ITGA11 | 1740112 6590110 | 5751 | 0.012 | 0.4616 | No | ||
| 59 | GPR37 | 2320093 | 5814 | 0.012 | 0.4587 | No | ||
| 60 | DMD | 1740041 3990332 | 5912 | 0.012 | 0.4539 | No | ||
| 61 | ITGA6 | 3830129 | 6257 | 0.010 | 0.4357 | No | ||
| 62 | CAMK2A | 1740333 1940112 4150292 | 6487 | 0.009 | 0.4236 | No | ||
| 63 | SORCS2 | 6350520 | 6635 | 0.008 | 0.4160 | No | ||
| 64 | DSCR1L1 | 940438 | 6649 | 0.008 | 0.4156 | No | ||
| 65 | MT3 | 1450537 | 6654 | 0.008 | 0.4157 | No | ||
| 66 | GNB3 | 1230167 | 6878 | 0.007 | 0.4039 | No | ||
| 67 | CHRND | 840403 2260670 | 7045 | 0.007 | 0.3952 | No | ||
| 68 | AHI1 | 460520 | 7181 | 0.006 | 0.3881 | No | ||
| 69 | TRPV3 | 540341 | 7652 | 0.005 | 0.3628 | No | ||
| 70 | ERBB3 | 5890372 | 7825 | 0.004 | 0.3537 | No | ||
| 71 | TACR1 | 70358 3840411 | 8066 | 0.003 | 0.3408 | No | ||
| 72 | PLEC1 | 6520292 50500 380075 1570088 2510692 2630497 2680017 2900746 4560497 4850376 6290398 7040471 | 8100 | 0.003 | 0.3391 | No | ||
| 73 | NR4A1 | 6290161 | 8225 | 0.003 | 0.3325 | No | ||
| 74 | CLSTN2 | 6900546 | 8239 | 0.003 | 0.3320 | No | ||
| 75 | KCNIP2 | 60088 1780324 | 8263 | 0.003 | 0.3308 | No | ||
| 76 | FSCN2 | 1230039 | 8317 | 0.003 | 0.3281 | No | ||
| 77 | PRDM16 | 4120541 | 8361 | 0.003 | 0.3258 | No | ||
| 78 | CALB1 | 460070 | 8880 | 0.001 | 0.2978 | No | ||
| 79 | POU4F3 | 2690035 | 8957 | 0.001 | 0.2938 | No | ||
| 80 | SEZ6 | 3450722 | 9003 | 0.001 | 0.2913 | No | ||
| 81 | PPARA | 2060026 | 9022 | 0.001 | 0.2904 | No | ||
| 82 | NDRG4 | 6510685 | 9446 | -0.000 | 0.2675 | No | ||
| 83 | SFTPC | 3290133 | 9508 | -0.000 | 0.2642 | No | ||
| 84 | APBB1 | 2690338 | 9604 | -0.001 | 0.2591 | No | ||
| 85 | PITX3 | 3130162 | 9608 | -0.001 | 0.2589 | No | ||
| 86 | NHLH1 | 6100452 | 9737 | -0.001 | 0.2520 | No | ||
| 87 | KCND3 | 670041 | 9966 | -0.002 | 0.2398 | No | ||
| 88 | THBS3 | 2360465 4050176 | 10081 | -0.002 | 0.2336 | No | ||
| 89 | KIF9 | 3710739 | 10203 | -0.002 | 0.2272 | No | ||
| 90 | IRX6 | 4590025 | 10210 | -0.002 | 0.2269 | No | ||
| 91 | ELA3B | 3990711 4060731 | 10228 | -0.002 | 0.2261 | No | ||
| 92 | PRPH | 2060136 | 10394 | -0.003 | 0.2173 | No | ||
| 93 | HOXC12 | 5270411 | 10430 | -0.003 | 0.2155 | No | ||
| 94 | KCNQ4 | 3800438 7050400 | 10503 | -0.003 | 0.2117 | No | ||
| 95 | RAB26 | 5910025 | 10510 | -0.003 | 0.2115 | No | ||
| 96 | ARHGEF12 | 3990195 | 10568 | -0.003 | 0.2086 | No | ||
| 97 | CYP1A1 | 4230113 | 10676 | -0.004 | 0.2029 | No | ||
| 98 | KIF5B | 770397 5420168 | 10698 | -0.004 | 0.2019 | No | ||
| 99 | ARL4A | 6290091 | 10711 | -0.004 | 0.2014 | No | ||
| 100 | SLC17A3 | 1780463 5670435 | 10745 | -0.004 | 0.1998 | No | ||
| 101 | SEMA3F | 5420601 6770731 | 10857 | -0.004 | 0.1939 | No | ||
| 102 | KCNJ2 | 630019 | 11058 | -0.005 | 0.1833 | No | ||
| 103 | ACTA1 | 840538 | 11213 | -0.005 | 0.1751 | No | ||
| 104 | KCNA4 | 2320706 | 11304 | -0.005 | 0.1705 | No | ||
| 105 | IRX5 | 2630494 | 11568 | -0.006 | 0.1565 | No | ||
| 106 | SOST | 1170195 | 11698 | -0.007 | 0.1498 | No | ||
| 107 | ELAVL3 | 2850014 | 11746 | -0.007 | 0.1475 | No | ||
| 108 | ITPR2 | 5360128 | 11774 | -0.007 | 0.1464 | No | ||
| 109 | TREX2 | 4920707 | 12028 | -0.008 | 0.1330 | No | ||
| 110 | FGF11 | 840292 | 12253 | -0.009 | 0.1212 | No | ||
| 111 | ANKRD2 | 6590427 | 12260 | -0.009 | 0.1212 | No | ||
| 112 | CYP26A1 | 380022 2690600 | 12274 | -0.009 | 0.1209 | No | ||
| 113 | CASKIN2 | 2030113 | 12279 | -0.009 | 0.1211 | No | ||
| 114 | CPNE1 | 3440131 | 12619 | -0.011 | 0.1031 | No | ||
| 115 | FZD5 | 4070452 | 12697 | -0.011 | 0.0994 | No | ||
| 116 | BDNF | 2940128 3520368 | 12799 | -0.012 | 0.0944 | No | ||
| 117 | UBTF | 4210450 6840403 | 13079 | -0.014 | 0.0799 | No | ||
| 118 | RNF128 | 1230300 5670273 | 13141 | -0.014 | 0.0771 | No | ||
| 119 | CDH2 | 520435 2450451 2760025 6650477 | 13151 | -0.014 | 0.0772 | No | ||
| 120 | PRSS12 | 1230364 6220059 | 13198 | -0.015 | 0.0753 | No | ||
| 121 | HRH3 | 6180040 | 13286 | -0.015 | 0.0712 | No | ||
| 122 | WDR20 | 1240373 | 13325 | -0.016 | 0.0698 | No | ||
| 123 | PAFAH1B1 | 4230333 6420121 6450066 | 13768 | -0.020 | 0.0467 | No | ||
| 124 | PRKAG1 | 6450091 | 14134 | -0.024 | 0.0278 | No | ||
| 125 | LBX1 | 1300164 4060204 | 14140 | -0.024 | 0.0285 | No | ||
| 126 | SLC39A5 | 2900100 4610440 | 14404 | -0.028 | 0.0154 | No | ||
| 127 | ARRDC3 | 4610390 | 14417 | -0.028 | 0.0159 | No | ||
| 128 | JMJD1C | 940575 2120025 | 14568 | -0.031 | 0.0090 | No | ||
| 129 | SPAG9 | 580497 2510180 4210685 | 14622 | -0.032 | 0.0074 | No | ||
| 130 | EPHA2 | 5890056 | 14688 | -0.033 | 0.0052 | No | ||
| 131 | GRIK3 | 6380592 | 14790 | -0.035 | 0.0012 | No | ||
| 132 | EIF2C1 | 6900551 | 14847 | -0.036 | -0.0004 | No | ||
| 133 | FHOD1 | 2680279 | 14888 | -0.037 | -0.0011 | No | ||
| 134 | FHL3 | 1090369 2650017 | 14980 | -0.040 | -0.0044 | No | ||
| 135 | ZIC4 | 1500082 | 15082 | -0.042 | -0.0082 | No | ||
| 136 | ACOX3 | 1580292 3390239 | 15291 | -0.049 | -0.0175 | No | ||
| 137 | PACSIN3 | 4540369 4540400 5360037 6940129 | 15454 | -0.055 | -0.0240 | No | ||
| 138 | SELL | 1190500 | 15592 | -0.060 | -0.0290 | No | ||
| 139 | WDTC1 | 1690541 | 15665 | -0.063 | -0.0304 | No | ||
| 140 | VDP | 2320403 | 15772 | -0.068 | -0.0334 | No | ||
| 141 | CYLD | 6590079 | 15802 | -0.069 | -0.0322 | No | ||
| 142 | REL | 360707 | 15811 | -0.069 | -0.0298 | No | ||
| 143 | POFUT1 | 1570458 | 15815 | -0.070 | -0.0272 | No | ||
| 144 | DCLRE1C | 1450041 2360091 3290671 3390739 | 15872 | -0.073 | -0.0273 | No | ||
| 145 | YARS | 70739 | 16063 | -0.083 | -0.0342 | No | ||
| 146 | TAZ | 7100193 | 16109 | -0.086 | -0.0332 | No | ||
| 147 | PLAGL2 | 1770148 | 16329 | -0.099 | -0.0411 | No | ||
| 148 | SIPA1 | 5220687 | 16402 | -0.105 | -0.0408 | No | ||
| 149 | CENTG3 | 2470079 4610092 5050170 | 16528 | -0.115 | -0.0429 | No | ||
| 150 | DNAJC11 | 360465 3390215 | 16777 | -0.135 | -0.0509 | No | ||
| 151 | POLL | 5910673 6350672 | 16961 | -0.153 | -0.0546 | No | ||
| 152 | MTX1 | 5910059 6550181 | 16967 | -0.154 | -0.0487 | No | ||
| 153 | BCL7A | 4560095 | 17170 | -0.178 | -0.0524 | No | ||
| 154 | FMNL1 | 5860347 | 17321 | -0.207 | -0.0522 | No | ||
| 155 | HMGN2 | 3140091 | 17469 | -0.232 | -0.0509 | No | ||
| 156 | SH3GLB2 | 2030086 | 17498 | -0.238 | -0.0428 | No | ||
| 157 | PSIP1 | 1780082 3190435 5050594 | 17554 | -0.248 | -0.0358 | No | ||
| 158 | CKM | 1450524 | 17637 | -0.267 | -0.0295 | No | ||
| 159 | CACNA1H | 1230279 | 17639 | -0.267 | -0.0187 | No | ||
| 160 | FBS1 | 2570520 | 17946 | -0.364 | -0.0206 | No | ||
| 161 | HMGA1 | 6580408 | 18028 | -0.394 | -0.0091 | No | ||
| 162 | CFL1 | 2340735 | 18071 | -0.410 | 0.0051 | No | ||
| 163 | LEF1 | 2470082 7100288 | 18318 | -0.604 | 0.0161 | No |