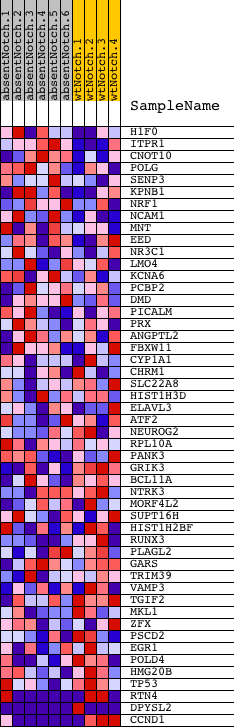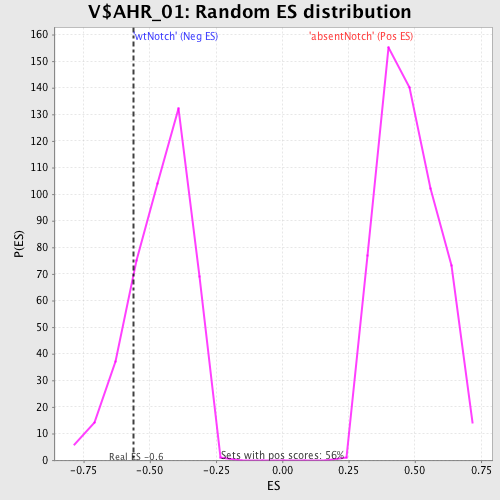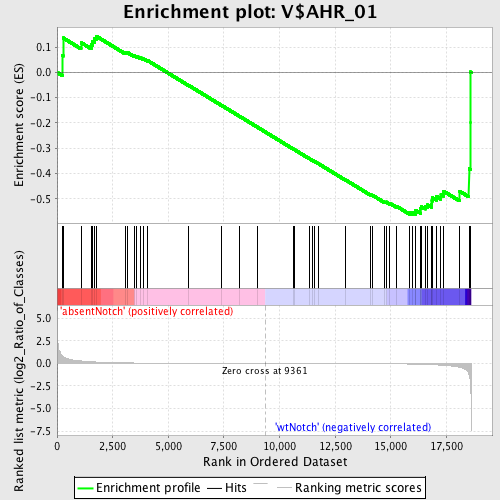
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | V$AHR_01 |
| Enrichment Score (ES) | -0.56226003 |
| Normalized Enrichment Score (NES) | -1.2249964 |
| Nominal p-value | 0.17579909 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | H1F0 | 1400408 | 230 | 0.849 | 0.0668 | No | ||
| 2 | ITPR1 | 3450519 | 269 | 0.768 | 0.1364 | No | ||
| 3 | CNOT10 | 2690541 | 1075 | 0.266 | 0.1179 | No | ||
| 4 | POLG | 780465 | 1534 | 0.181 | 0.1101 | No | ||
| 5 | SENP3 | 4200082 | 1581 | 0.174 | 0.1238 | No | ||
| 6 | KPNB1 | 1690138 | 1675 | 0.160 | 0.1338 | No | ||
| 7 | NRF1 | 2650195 | 1758 | 0.152 | 0.1435 | No | ||
| 8 | NCAM1 | 3140026 | 3059 | 0.058 | 0.0789 | No | ||
| 9 | MNT | 4920575 | 3146 | 0.055 | 0.0794 | No | ||
| 10 | EED | 2320373 | 3459 | 0.043 | 0.0666 | No | ||
| 11 | NR3C1 | 630563 | 3575 | 0.040 | 0.0642 | No | ||
| 12 | LMO4 | 3800746 | 3754 | 0.036 | 0.0579 | No | ||
| 13 | KCNA6 | 450291 1400086 1660291 2470576 | 3895 | 0.032 | 0.0534 | No | ||
| 14 | PCBP2 | 5690048 | 4080 | 0.029 | 0.0462 | No | ||
| 15 | DMD | 1740041 3990332 | 5912 | 0.012 | -0.0513 | No | ||
| 16 | PICALM | 1940280 | 7377 | 0.005 | -0.1296 | No | ||
| 17 | PRX | 2260400 4200347 | 8209 | 0.003 | -0.1741 | No | ||
| 18 | ANGPTL2 | 6980152 6760162 | 9026 | 0.001 | -0.2180 | No | ||
| 19 | FBXW11 | 6450632 | 10645 | -0.003 | -0.3048 | No | ||
| 20 | CYP1A1 | 4230113 | 10676 | -0.004 | -0.3061 | No | ||
| 21 | CHRM1 | 4280619 | 11355 | -0.006 | -0.3421 | No | ||
| 22 | SLC22A8 | 3520204 | 11482 | -0.006 | -0.3483 | No | ||
| 23 | HIST1H3D | 70687 | 11569 | -0.006 | -0.3523 | No | ||
| 24 | ELAVL3 | 2850014 | 11746 | -0.007 | -0.3611 | No | ||
| 25 | ATF2 | 2260348 2480441 | 12975 | -0.013 | -0.4261 | No | ||
| 26 | NEUROG2 | 5420278 | 14082 | -0.023 | -0.4835 | No | ||
| 27 | RPL10A | 4850066 | 14172 | -0.024 | -0.4860 | No | ||
| 28 | PANK3 | 3060687 | 14733 | -0.034 | -0.5130 | No | ||
| 29 | GRIK3 | 6380592 | 14790 | -0.035 | -0.5127 | No | ||
| 30 | BCL11A | 6860369 | 14956 | -0.039 | -0.5180 | No | ||
| 31 | NTRK3 | 4050687 4070167 4610026 6100484 | 15238 | -0.047 | -0.5287 | No | ||
| 32 | MORF4L2 | 6450133 | 15852 | -0.072 | -0.5550 | Yes | ||
| 33 | SUPT16H | 1240333 | 15987 | -0.078 | -0.5550 | Yes | ||
| 34 | HIST1H2BF | 5420402 | 16099 | -0.085 | -0.5530 | Yes | ||
| 35 | RUNX3 | 3800546 | 16110 | -0.086 | -0.5455 | Yes | ||
| 36 | PLAGL2 | 1770148 | 16329 | -0.099 | -0.5480 | Yes | ||
| 37 | GARS | 1230601 | 16330 | -0.099 | -0.5388 | Yes | ||
| 38 | TRIM39 | 6550441 | 16363 | -0.102 | -0.5310 | Yes | ||
| 39 | VAMP3 | 7100050 | 16543 | -0.116 | -0.5298 | Yes | ||
| 40 | TGIF2 | 3840364 4050520 | 16656 | -0.123 | -0.5244 | Yes | ||
| 41 | MKL1 | 6840161 | 16835 | -0.139 | -0.5210 | Yes | ||
| 42 | ZFX | 5900400 | 16848 | -0.141 | -0.5085 | Yes | ||
| 43 | PSCD2 | 3870113 | 16860 | -0.142 | -0.4958 | Yes | ||
| 44 | EGR1 | 4610347 | 17040 | -0.161 | -0.4904 | Yes | ||
| 45 | POLD4 | 2320082 | 17254 | -0.194 | -0.4838 | Yes | ||
| 46 | HMG20B | 2450465 | 17361 | -0.214 | -0.4696 | Yes | ||
| 47 | TP53 | 6130707 | 18087 | -0.415 | -0.4699 | Yes | ||
| 48 | RTN4 | 50725 630647 870097 870184 | 18513 | -1.196 | -0.3812 | Yes | ||
| 49 | DPYSL2 | 2100427 3130112 5700324 | 18580 | -2.012 | -0.1970 | Yes | ||
| 50 | CCND1 | 460524 770309 3120576 6980398 | 18583 | -2.132 | 0.0018 | Yes |