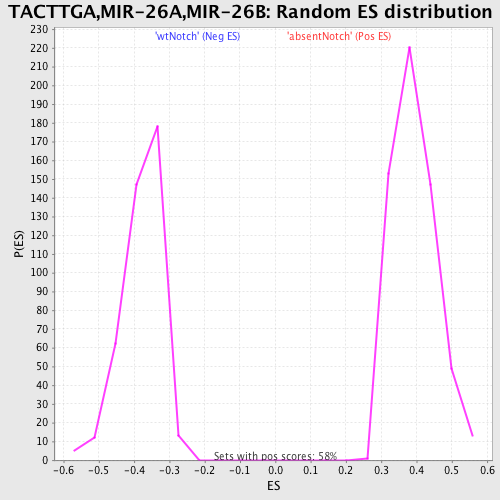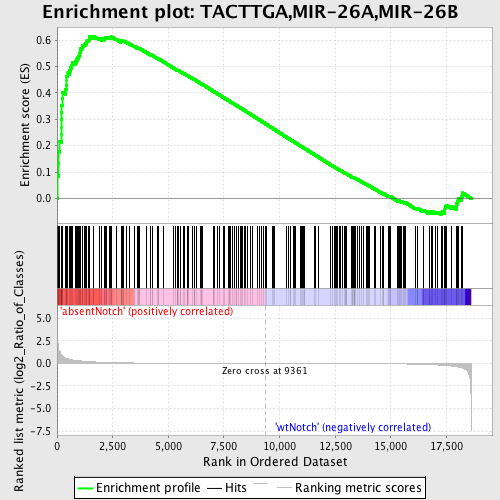
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | TACTTGA,MIR-26A,MIR-26B |
| Enrichment Score (ES) | 0.6150667 |
| Normalized Enrichment Score (NES) | 1.5639756 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.24174252 |
| FWER p-Value | 0.775 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARHGAP17 | 4810095 5360563 5900427 | 21 | 2.860 | 0.0866 | Yes | ||
| 2 | CCND2 | 5340167 | 59 | 1.612 | 0.1341 | Yes | ||
| 3 | SLC38A2 | 1470242 3800026 | 81 | 1.464 | 0.1779 | Yes | ||
| 4 | VANGL2 | 70097 870075 | 116 | 1.348 | 0.2174 | Yes | ||
| 5 | EIF5 | 6380750 | 192 | 0.927 | 0.2417 | Yes | ||
| 6 | CMTM4 | 360600 3520332 | 195 | 0.922 | 0.2699 | Yes | ||
| 7 | UBE4B | 780008 3610154 | 197 | 0.919 | 0.2981 | Yes | ||
| 8 | ADM | 4810121 | 206 | 0.898 | 0.3252 | Yes | ||
| 9 | PPP3CB | 6020156 | 211 | 0.891 | 0.3523 | Yes | ||
| 10 | ENC1 | 4810348 | 228 | 0.859 | 0.3778 | Yes | ||
| 11 | INHBB | 2680092 | 240 | 0.832 | 0.4028 | Yes | ||
| 12 | SRPK1 | 450110 | 382 | 0.611 | 0.4138 | Yes | ||
| 13 | SH3KBP1 | 670102 2850458 7000309 | 419 | 0.571 | 0.4294 | Yes | ||
| 14 | CCNE1 | 110064 | 435 | 0.561 | 0.4458 | Yes | ||
| 15 | BHLHB2 | 7040603 | 437 | 0.560 | 0.4629 | Yes | ||
| 16 | PPP3R1 | 1190041 1570487 | 487 | 0.504 | 0.4758 | Yes | ||
| 17 | USP3 | 2060332 | 575 | 0.440 | 0.4845 | Yes | ||
| 18 | YPEL1 | 6590435 | 611 | 0.424 | 0.4956 | Yes | ||
| 19 | MAPK6 | 6760520 | 664 | 0.398 | 0.5050 | Yes | ||
| 20 | MOBK1B | 4810373 | 702 | 0.379 | 0.5146 | Yes | ||
| 21 | RNF6 | 2640035 | 814 | 0.337 | 0.5190 | Yes | ||
| 22 | EZH2 | 6130605 6380524 | 873 | 0.321 | 0.5256 | Yes | ||
| 23 | TOB1 | 4150138 | 922 | 0.306 | 0.5324 | Yes | ||
| 24 | EIF4G2 | 3800575 6860184 | 959 | 0.293 | 0.5395 | Yes | ||
| 25 | SLC7A11 | 2850138 | 1013 | 0.280 | 0.5452 | Yes | ||
| 26 | TRIB2 | 4120605 | 1027 | 0.277 | 0.5530 | Yes | ||
| 27 | TTC13 | 3520750 | 1045 | 0.274 | 0.5605 | Yes | ||
| 28 | TMEM33 | 2340022 3940750 | 1057 | 0.271 | 0.5682 | Yes | ||
| 29 | CBFB | 1850551 2650035 | 1121 | 0.257 | 0.5727 | Yes | ||
| 30 | CD200 | 940725 5260647 | 1136 | 0.253 | 0.5797 | Yes | ||
| 31 | TAF12 | 3710746 4920300 6180603 | 1208 | 0.240 | 0.5832 | Yes | ||
| 32 | OSBPL2 | 5690215 | 1290 | 0.223 | 0.5856 | Yes | ||
| 33 | CUGBP2 | 6180121 6840139 | 1302 | 0.222 | 0.5918 | Yes | ||
| 34 | EP400 | 3940722 | 1336 | 0.217 | 0.5967 | Yes | ||
| 35 | GSK3B | 5360348 | 1401 | 0.203 | 0.5994 | Yes | ||
| 36 | MCL1 | 1660672 | 1437 | 0.196 | 0.6036 | Yes | ||
| 37 | SLC4A7 | 2370056 | 1445 | 0.195 | 0.6092 | Yes | ||
| 38 | SMAD4 | 5670519 | 1447 | 0.194 | 0.6151 | Yes | ||
| 39 | UBE2G1 | 2230546 5420528 7040463 | 1623 | 0.167 | 0.6107 | No | ||
| 40 | ARL4C | 1190279 | 1655 | 0.163 | 0.6140 | No | ||
| 41 | PHTF2 | 6590053 | 1887 | 0.138 | 0.6057 | No | ||
| 42 | GTF3C2 | 670494 | 2002 | 0.125 | 0.6033 | No | ||
| 43 | IGF1 | 1990193 3130377 3290280 | 2010 | 0.125 | 0.6068 | No | ||
| 44 | PIM1 | 630047 | 2131 | 0.115 | 0.6038 | No | ||
| 45 | CDK6 | 4920253 | 2151 | 0.113 | 0.6062 | No | ||
| 46 | RTF1 | 2100735 | 2153 | 0.113 | 0.6096 | No | ||
| 47 | KPNA2 | 3450114 | 2206 | 0.108 | 0.6101 | No | ||
| 48 | SLC12A2 | 4610019 6040672 | 2239 | 0.106 | 0.6116 | No | ||
| 49 | RAP2C | 1690132 | 2333 | 0.099 | 0.6096 | No | ||
| 50 | BCR | 2260020 4230180 6040195 | 2404 | 0.094 | 0.6087 | No | ||
| 51 | SH3D19 | 6020520 | 2434 | 0.093 | 0.6099 | No | ||
| 52 | MAN2A1 | 6650176 | 2450 | 0.092 | 0.6119 | No | ||
| 53 | CHFR | 3390050 6450300 6450736 | 2647 | 0.078 | 0.6037 | No | ||
| 54 | PAK2 | 360438 7050068 | 2871 | 0.066 | 0.5936 | No | ||
| 55 | MKNK2 | 1240020 5270092 | 2879 | 0.066 | 0.5952 | No | ||
| 56 | HPGD | 6770192 | 2886 | 0.066 | 0.5969 | No | ||
| 57 | NRIP1 | 3190039 | 2944 | 0.063 | 0.5958 | No | ||
| 58 | SRP19 | 780450 3120075 | 2976 | 0.062 | 0.5960 | No | ||
| 59 | PTPN13 | 2060681 | 2998 | 0.061 | 0.5967 | No | ||
| 60 | THRAP2 | 4920600 | 3114 | 0.056 | 0.5922 | No | ||
| 61 | NEK6 | 3360687 | 3246 | 0.050 | 0.5866 | No | ||
| 62 | STK39 | 50692 | 3472 | 0.043 | 0.5757 | No | ||
| 63 | USP37 | 2470433 | 3592 | 0.039 | 0.5704 | No | ||
| 64 | SPOCK2 | 6650369 | 3638 | 0.038 | 0.5692 | No | ||
| 65 | PELI2 | 1940364 | 3665 | 0.038 | 0.5689 | No | ||
| 66 | MARK1 | 450484 | 3714 | 0.037 | 0.5674 | No | ||
| 67 | PDGFRA | 2940332 | 3717 | 0.037 | 0.5684 | No | ||
| 68 | SLC1A1 | 520372 6420059 | 3997 | 0.031 | 0.5542 | No | ||
| 69 | VAMP1 | 450735 1940707 | 4196 | 0.027 | 0.5443 | No | ||
| 70 | ARID2 | 110647 5390435 | 4218 | 0.027 | 0.5440 | No | ||
| 71 | ACADSB | 520170 | 4291 | 0.026 | 0.5409 | No | ||
| 72 | PCDH9 | 2100136 | 4300 | 0.026 | 0.5412 | No | ||
| 73 | PALMD | 2900369 | 4498 | 0.023 | 0.5312 | No | ||
| 74 | PRKCD | 770592 | 4555 | 0.022 | 0.5289 | No | ||
| 75 | STAC2 | 7100022 | 4560 | 0.022 | 0.5293 | No | ||
| 76 | DLG5 | 450215 | 4764 | 0.020 | 0.5189 | No | ||
| 77 | MAB21L1 | 1450634 1580092 | 5226 | 0.016 | 0.4944 | No | ||
| 78 | MIB1 | 3800537 | 5336 | 0.015 | 0.4889 | No | ||
| 79 | FANCA | 6130070 | 5405 | 0.015 | 0.4856 | No | ||
| 80 | PITPNC1 | 3990017 | 5424 | 0.015 | 0.4851 | No | ||
| 81 | TBC1D4 | 6420397 | 5430 | 0.014 | 0.4853 | No | ||
| 82 | ACSL3 | 3140195 | 5465 | 0.014 | 0.4839 | No | ||
| 83 | HTR2A | 360037 6330255 | 5546 | 0.014 | 0.4799 | No | ||
| 84 | RGS4 | 2970711 | 5562 | 0.014 | 0.4795 | No | ||
| 85 | SENP5 | 4560537 6760465 | 5702 | 0.013 | 0.4724 | No | ||
| 86 | RTN1 | 4150452 4850176 | 5728 | 0.013 | 0.4714 | No | ||
| 87 | ITGA5 | 5550520 | 5859 | 0.012 | 0.4647 | No | ||
| 88 | OSBPL11 | 1770044 | 5869 | 0.012 | 0.4646 | No | ||
| 89 | LGR4 | 1660309 | 5872 | 0.012 | 0.4648 | No | ||
| 90 | HAO1 | 7050093 | 5913 | 0.012 | 0.4630 | No | ||
| 91 | LIF | 1410373 2060129 3990338 | 6105 | 0.010 | 0.4530 | No | ||
| 92 | RAP1A | 1090025 | 6160 | 0.010 | 0.4503 | No | ||
| 93 | VLDLR | 870722 3060047 5340452 6550131 | 6164 | 0.010 | 0.4505 | No | ||
| 94 | PTP4A1 | 130692 | 6189 | 0.010 | 0.4495 | No | ||
| 95 | NUP50 | 1240519 2640390 2650471 4730133 | 6248 | 0.010 | 0.4466 | No | ||
| 96 | COL10A1 | 2360307 | 6455 | 0.009 | 0.4357 | No | ||
| 97 | KCNH7 | 7510068 | 6472 | 0.009 | 0.4351 | No | ||
| 98 | E2F7 | 4200056 | 6494 | 0.009 | 0.4342 | No | ||
| 99 | SSX2IP | 4610176 | 6515 | 0.009 | 0.4334 | No | ||
| 100 | SMAD1 | 630537 1850333 | 7014 | 0.007 | 0.4066 | No | ||
| 101 | ASPN | 2450021 | 7017 | 0.007 | 0.4067 | No | ||
| 102 | FGD1 | 4780021 | 7066 | 0.006 | 0.4042 | No | ||
| 103 | LRRC16 | 360022 6450070 | 7202 | 0.006 | 0.3971 | No | ||
| 104 | SRGAP1 | 3940253 | 7317 | 0.006 | 0.3911 | No | ||
| 105 | PNRC1 | 360025 | 7460 | 0.005 | 0.3835 | No | ||
| 106 | ACVR1C | 5890044 | 7503 | 0.005 | 0.3814 | No | ||
| 107 | YTHDF3 | 4540632 4670717 7040609 | 7684 | 0.005 | 0.3717 | No | ||
| 108 | JAG1 | 3440390 | 7728 | 0.004 | 0.3695 | No | ||
| 109 | STYX | 1090358 | 7766 | 0.004 | 0.3677 | No | ||
| 110 | DCDC2 | 7000338 | 7805 | 0.004 | 0.3657 | No | ||
| 111 | RCN2 | 840324 | 7863 | 0.004 | 0.3627 | No | ||
| 112 | UBE2D1 | 1850400 | 7957 | 0.004 | 0.3578 | No | ||
| 113 | GABRB3 | 4150164 | 8053 | 0.004 | 0.3527 | No | ||
| 114 | BTBD7 | 730288 | 8055 | 0.003 | 0.3528 | No | ||
| 115 | NFE2L3 | 780100 | 8169 | 0.003 | 0.3468 | No | ||
| 116 | NTN4 | 6940398 | 8243 | 0.003 | 0.3429 | No | ||
| 117 | PHF6 | 3120632 3990041 | 8309 | 0.003 | 0.3394 | No | ||
| 118 | PLEKHH1 | 4780280 | 8316 | 0.003 | 0.3392 | No | ||
| 119 | PCDH18 | 430010 | 8434 | 0.002 | 0.3329 | No | ||
| 120 | SYT10 | 5550048 | 8457 | 0.002 | 0.3318 | No | ||
| 121 | USP9X | 130435 2370735 3120338 | 8579 | 0.002 | 0.3253 | No | ||
| 122 | SLC25A16 | 4200341 | 8699 | 0.002 | 0.3189 | No | ||
| 123 | STRBP | 4210594 5360239 | 8796 | 0.001 | 0.3137 | No | ||
| 124 | PAWR | 5130546 6330706 | 8986 | 0.001 | 0.3034 | No | ||
| 125 | CLASP2 | 2510139 | 9106 | 0.001 | 0.2970 | No | ||
| 126 | ATP1A2 | 110278 | 9164 | 0.001 | 0.2939 | No | ||
| 127 | PHLDB2 | 6590685 | 9295 | 0.000 | 0.2868 | No | ||
| 128 | ZDHHC6 | 510600 | 9378 | -0.000 | 0.2824 | No | ||
| 129 | MITF | 380056 | 9394 | -0.000 | 0.2816 | No | ||
| 130 | EIF3S10 | 1190079 3840176 | 9660 | -0.001 | 0.2672 | No | ||
| 131 | NOL4 | 5050446 5360519 | 9745 | -0.001 | 0.2626 | No | ||
| 132 | PTGS2 | 2510301 3170369 | 9763 | -0.001 | 0.2618 | No | ||
| 133 | MMP16 | 2680139 | 10299 | -0.002 | 0.2327 | No | ||
| 134 | PDHX | 870315 | 10398 | -0.003 | 0.2275 | No | ||
| 135 | PAPD4 | 1580673 730605 5270112 | 10468 | -0.003 | 0.2238 | No | ||
| 136 | KCNQ4 | 3800438 7050400 | 10503 | -0.003 | 0.2221 | No | ||
| 137 | TRPC3 | 840064 | 10604 | -0.003 | 0.2168 | No | ||
| 138 | RCN1 | 2480041 | 10657 | -0.003 | 0.2140 | No | ||
| 139 | NAP1L5 | 360576 | 10705 | -0.004 | 0.2116 | No | ||
| 140 | POF1B | 3800670 | 10954 | -0.004 | 0.1983 | No | ||
| 141 | SSH2 | 360056 | 10991 | -0.005 | 0.1964 | No | ||
| 142 | ZNF462 | 6900100 | 11026 | -0.005 | 0.1947 | No | ||
| 143 | KCNJ2 | 630019 | 11058 | -0.005 | 0.1932 | No | ||
| 144 | SLC24A4 | 2100487 | 11105 | -0.005 | 0.1908 | No | ||
| 145 | HOXA5 | 6840026 | 11121 | -0.005 | 0.1902 | No | ||
| 146 | KLF10 | 4850056 | 11559 | -0.006 | 0.1666 | No | ||
| 147 | KCNK1 | 1050068 | 11617 | -0.007 | 0.1637 | No | ||
| 148 | RPGR | 5910711 | 11733 | -0.007 | 0.1577 | No | ||
| 149 | ATP11C | 770672 7040129 | 12282 | -0.009 | 0.1282 | No | ||
| 150 | PURA | 3870156 | 12399 | -0.010 | 0.1222 | No | ||
| 151 | HGF | 3360593 | 12476 | -0.010 | 0.1184 | No | ||
| 152 | EPC2 | 2470095 | 12499 | -0.010 | 0.1175 | No | ||
| 153 | ACBD5 | 3440397 6020600 | 12559 | -0.011 | 0.1146 | No | ||
| 154 | MTX2 | 6660288 | 12596 | -0.011 | 0.1130 | No | ||
| 155 | RANBP10 | 2340184 | 12671 | -0.011 | 0.1093 | No | ||
| 156 | ATPAF1 | 2470164 | 12740 | -0.012 | 0.1059 | No | ||
| 157 | PLCB1 | 2570050 7100102 | 12828 | -0.012 | 0.1016 | No | ||
| 158 | HAS3 | 4200692 | 12899 | -0.013 | 0.0982 | No | ||
| 159 | USP25 | 3520035 | 12961 | -0.013 | 0.0953 | No | ||
| 160 | ATF2 | 2260348 2480441 | 12975 | -0.013 | 0.0950 | No | ||
| 161 | AMPH | 2260338 3140576 | 12988 | -0.013 | 0.0947 | No | ||
| 162 | WNK3 | 4060053 | 13023 | -0.013 | 0.0933 | No | ||
| 163 | ALS2CR2 | 6450128 7100092 | 13249 | -0.015 | 0.0815 | No | ||
| 164 | CDK2AP1 | 2340156 | 13292 | -0.015 | 0.0797 | No | ||
| 165 | LTBP1 | 780035 2100746 2760402 3780551 6550184 6860193 | 13296 | -0.015 | 0.0800 | No | ||
| 166 | PBEF1 | 1090278 | 13336 | -0.016 | 0.0784 | No | ||
| 167 | ZFHX4 | 6110167 | 13345 | -0.016 | 0.0784 | No | ||
| 168 | CSNK1G1 | 840082 1230575 3940647 | 13361 | -0.016 | 0.0781 | No | ||
| 169 | USP15 | 610592 3520504 | 13377 | -0.016 | 0.0778 | No | ||
| 170 | SLC4A4 | 1780075 1850040 4810068 | 13398 | -0.016 | 0.0772 | No | ||
| 171 | RKHD3 | 6180471 | 13499 | -0.017 | 0.0723 | No | ||
| 172 | DAPK1 | 5570010 | 13577 | -0.018 | 0.0686 | No | ||
| 173 | COL19A1 | 6220441 | 13680 | -0.019 | 0.0637 | No | ||
| 174 | PLOD2 | 3360427 | 13786 | -0.020 | 0.0586 | No | ||
| 175 | PCK1 | 7000358 | 13885 | -0.021 | 0.0539 | No | ||
| 176 | CAMSAP1 | 1580131 2120494 6620446 | 13930 | -0.021 | 0.0521 | No | ||
| 177 | ATM | 3610110 4050524 | 14018 | -0.022 | 0.0481 | No | ||
| 178 | NAB1 | 430739 1230114 3390736 6660056 | 14042 | -0.023 | 0.0476 | No | ||
| 179 | MXI1 | 5050064 5130484 | 14268 | -0.026 | 0.0361 | No | ||
| 180 | HMGA2 | 2940121 3390647 5130279 6400136 | 14320 | -0.027 | 0.0342 | No | ||
| 181 | LSM11 | 2350167 | 14536 | -0.030 | 0.0234 | No | ||
| 182 | XKRX | 3800192 | 14642 | -0.032 | 0.0187 | No | ||
| 183 | EPHA2 | 5890056 | 14688 | -0.033 | 0.0173 | No | ||
| 184 | AMOT | 1940121 2940647 6590093 | 14691 | -0.033 | 0.0182 | No | ||
| 185 | PHF21A | 610014 | 14916 | -0.038 | 0.0071 | No | ||
| 186 | SALL1 | 5420020 7050195 | 14926 | -0.038 | 0.0078 | No | ||
| 187 | WNT5A | 840685 3120152 | 14967 | -0.039 | 0.0069 | No | ||
| 188 | CACNA1C | 1660128 3780300 3870092 6290403 6900301 | 14972 | -0.039 | 0.0079 | No | ||
| 189 | BAK1 | 6450563 | 15311 | -0.050 | -0.0090 | No | ||
| 190 | ALDH5A1 | 7040750 | 15330 | -0.050 | -0.0084 | No | ||
| 191 | APC | 3850484 5860722 | 15367 | -0.052 | -0.0088 | No | ||
| 192 | RPS6KA2 | 2970161 4730278 | 15449 | -0.055 | -0.0115 | No | ||
| 193 | ULK1 | 6100315 | 15490 | -0.056 | -0.0120 | No | ||
| 194 | AMMECR1 | 3800435 | 15575 | -0.059 | -0.0147 | No | ||
| 195 | ULK2 | 50041 5050176 | 15631 | -0.061 | -0.0158 | No | ||
| 196 | ST8SIA4 | 2680605 3060215 | 15659 | -0.063 | -0.0153 | No | ||
| 197 | CPSF2 | 6040709 | 16088 | -0.085 | -0.0360 | No | ||
| 198 | ARPP-19 | 5700025 | 16180 | -0.089 | -0.0382 | No | ||
| 199 | FA2H | 730672 | 16216 | -0.091 | -0.0373 | No | ||
| 200 | EHD1 | 3840731 | 16457 | -0.109 | -0.0470 | No | ||
| 201 | COX5A | 1450609 | 16459 | -0.110 | -0.0437 | No | ||
| 202 | NARG1 | 5910563 6350095 | 16732 | -0.131 | -0.0545 | No | ||
| 203 | TMEM2 | 1980037 3290020 | 16737 | -0.131 | -0.0507 | No | ||
| 204 | ZFX | 5900400 | 16848 | -0.141 | -0.0523 | No | ||
| 205 | MRAS | 2030524 | 16859 | -0.142 | -0.0485 | No | ||
| 206 | PFKFB3 | 630706 | 17014 | -0.158 | -0.0520 | No | ||
| 207 | BLOC1S2 | 1240215 | 17110 | -0.170 | -0.0520 | No | ||
| 208 | BRWD1 | 3130048 3140168 6400601 | 17267 | -0.197 | -0.0544 | No | ||
| 209 | ADAM19 | 5340075 | 17304 | -0.203 | -0.0501 | No | ||
| 210 | ABHD2 | 4850504 6840010 | 17429 | -0.226 | -0.0499 | No | ||
| 211 | NDFIP2 | 4070451 | 17433 | -0.226 | -0.0432 | No | ||
| 212 | ZDHHC18 | 6200059 | 17434 | -0.226 | -0.0362 | No | ||
| 213 | PTER | 3450685 | 17453 | -0.230 | -0.0301 | No | ||
| 214 | CELSR1 | 6510348 | 17483 | -0.234 | -0.0245 | No | ||
| 215 | FBXL19 | 4570039 | 17728 | -0.290 | -0.0289 | No | ||
| 216 | TNRC6A | 1240537 6130037 6200010 | 17948 | -0.365 | -0.0296 | No | ||
| 217 | DEPDC1B | 2120458 | 17960 | -0.367 | -0.0190 | No | ||
| 218 | POM121 | 70152 | 18001 | -0.384 | -0.0093 | No | ||
| 219 | HMGA1 | 6580408 | 18028 | -0.394 | 0.0013 | No | ||
| 220 | ALS2 | 3130546 6400070 | 18190 | -0.483 | 0.0074 | No | ||
| 221 | TMEM24 | 5860039 | 18227 | -0.512 | 0.0211 | No |