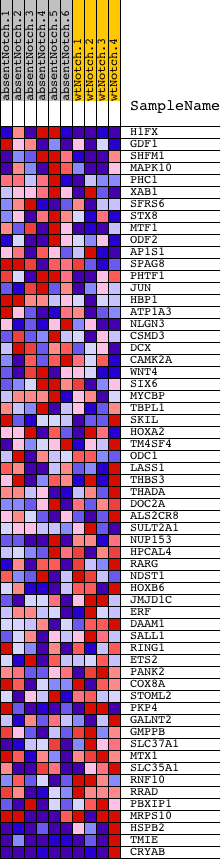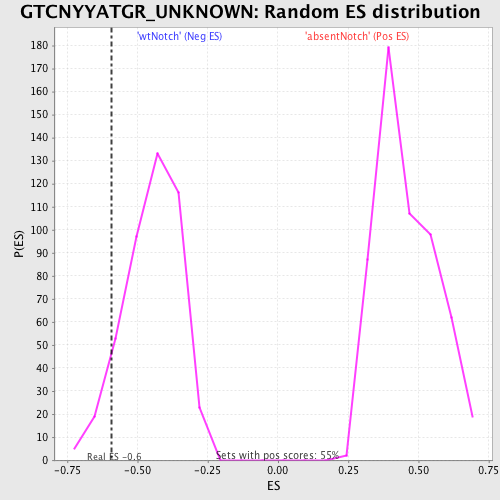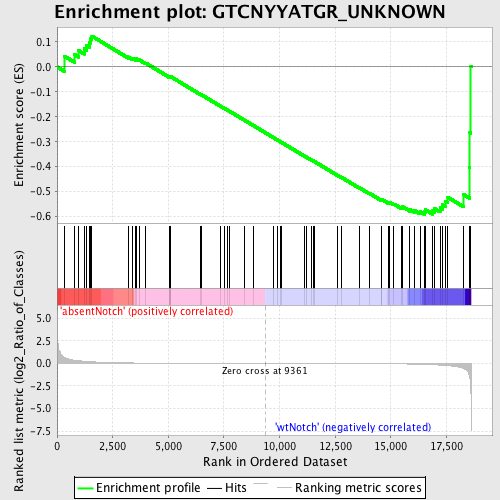
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | GTCNYYATGR_UNKNOWN |
| Enrichment Score (ES) | -0.5915244 |
| Normalized Enrichment Score (NES) | -1.3203725 |
| Nominal p-value | 0.07399103 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | H1FX | 1190088 | 317 | 0.683 | 0.0437 | No | ||
| 2 | GDF1 | 2350132 | 787 | 0.347 | 0.0493 | No | ||
| 3 | SHFM1 | 1570114 | 971 | 0.290 | 0.0652 | No | ||
| 4 | MAPK10 | 6110193 | 1224 | 0.237 | 0.0727 | No | ||
| 5 | PHC1 | 520195 | 1328 | 0.218 | 0.0865 | No | ||
| 6 | XAB1 | 3190671 | 1460 | 0.192 | 0.0965 | No | ||
| 7 | SFRS6 | 60224 | 1483 | 0.189 | 0.1121 | No | ||
| 8 | STX8 | 730411 3940427 6290471 | 1563 | 0.176 | 0.1235 | No | ||
| 9 | MTF1 | 520427 2340364 | 3201 | 0.052 | 0.0399 | No | ||
| 10 | ODF2 | 1660487 4150484 5690706 6940121 | 3383 | 0.045 | 0.0342 | No | ||
| 11 | AP1S1 | 2030471 4570520 6900603 | 3532 | 0.041 | 0.0299 | No | ||
| 12 | SPAG8 | 4280446 | 3552 | 0.041 | 0.0325 | No | ||
| 13 | PHTF1 | 7050441 | 3697 | 0.037 | 0.0280 | No | ||
| 14 | JUN | 840170 | 3990 | 0.031 | 0.0150 | No | ||
| 15 | HBP1 | 2510102 3130010 4210619 | 5045 | 0.017 | -0.0403 | No | ||
| 16 | ATP1A3 | 5690674 | 5061 | 0.017 | -0.0395 | No | ||
| 17 | NLGN3 | 2650463 | 5089 | 0.017 | -0.0395 | No | ||
| 18 | CSMD3 | 1660427 1940687 | 5105 | 0.017 | -0.0388 | No | ||
| 19 | DCX | 130075 5360025 7050673 | 6438 | 0.009 | -0.1098 | No | ||
| 20 | CAMK2A | 1740333 1940112 4150292 | 6487 | 0.009 | -0.1116 | No | ||
| 21 | WNT4 | 4150619 | 7335 | 0.006 | -0.1567 | No | ||
| 22 | SIX6 | 3140458 | 7502 | 0.005 | -0.1652 | No | ||
| 23 | MYCBP | 4280528 | 7657 | 0.005 | -0.1731 | No | ||
| 24 | TBPL1 | 3610332 | 7768 | 0.004 | -0.1786 | No | ||
| 25 | SKIL | 1090364 | 8417 | 0.003 | -0.2133 | No | ||
| 26 | HOXA2 | 2120121 | 8840 | 0.001 | -0.2360 | No | ||
| 27 | TM4SF4 | 540600 | 9742 | -0.001 | -0.2844 | No | ||
| 28 | ODC1 | 5670168 | 9886 | -0.001 | -0.2920 | No | ||
| 29 | LASS1 | 3440746 6110152 | 10052 | -0.002 | -0.3008 | No | ||
| 30 | THBS3 | 2360465 4050176 | 10081 | -0.002 | -0.3021 | No | ||
| 31 | THADA | 6760348 | 11118 | -0.005 | -0.3575 | No | ||
| 32 | DOC2A | 6020121 | 11191 | -0.005 | -0.3609 | No | ||
| 33 | ALS2CR8 | 4920088 | 11435 | -0.006 | -0.3735 | No | ||
| 34 | SULT2A1 | 2650286 | 11502 | -0.006 | -0.3765 | No | ||
| 35 | NUP153 | 7000452 | 11575 | -0.006 | -0.3798 | No | ||
| 36 | HPCAL4 | 2600484 | 12587 | -0.011 | -0.4333 | No | ||
| 37 | RARG | 6760136 | 12788 | -0.012 | -0.4431 | No | ||
| 38 | NDST1 | 1500427 5340121 | 13570 | -0.018 | -0.4836 | No | ||
| 39 | HOXB6 | 4210739 | 14022 | -0.023 | -0.5059 | No | ||
| 40 | JMJD1C | 940575 2120025 | 14568 | -0.031 | -0.5325 | No | ||
| 41 | ERF | 110551 1770278 | 14596 | -0.031 | -0.5312 | No | ||
| 42 | DAAM1 | 1500273 | 14899 | -0.037 | -0.5441 | No | ||
| 43 | SALL1 | 5420020 7050195 | 14926 | -0.038 | -0.5421 | No | ||
| 44 | RING1 | 4560180 | 15137 | -0.044 | -0.5495 | No | ||
| 45 | ETS2 | 360451 | 15465 | -0.055 | -0.5622 | No | ||
| 46 | PANK2 | 130446 | 15536 | -0.058 | -0.5609 | No | ||
| 47 | COX8A | 3780500 | 15855 | -0.072 | -0.5716 | No | ||
| 48 | STOML2 | 1940487 | 16052 | -0.082 | -0.5749 | No | ||
| 49 | PKP4 | 3710400 5890097 | 16321 | -0.099 | -0.5805 | No | ||
| 50 | GALNT2 | 2260279 4060239 | 16526 | -0.115 | -0.5813 | Yes | ||
| 51 | GMPPB | 4610538 | 16542 | -0.116 | -0.5719 | Yes | ||
| 52 | SLC37A1 | 5340050 | 16881 | -0.144 | -0.5773 | Yes | ||
| 53 | MTX1 | 5910059 6550181 | 16967 | -0.154 | -0.5682 | Yes | ||
| 54 | SLC35A1 | 2940750 3290364 | 17215 | -0.186 | -0.5650 | Yes | ||
| 55 | RNF10 | 1240438 | 17323 | -0.207 | -0.5523 | Yes | ||
| 56 | RRAD | 4280129 | 17442 | -0.228 | -0.5384 | Yes | ||
| 57 | PBXIP1 | 70131 | 17566 | -0.250 | -0.5228 | Yes | ||
| 58 | MRPS10 | 2060315 4880300 6040047 | 18268 | -0.549 | -0.5117 | Yes | ||
| 59 | HSPB2 | 1990110 | 18539 | -1.389 | -0.4028 | Yes | ||
| 60 | TMIE | 2370164 | 18552 | -1.580 | -0.2630 | Yes | ||
| 61 | CRYAB | 4810619 | 18600 | -2.995 | 0.0009 | Yes |