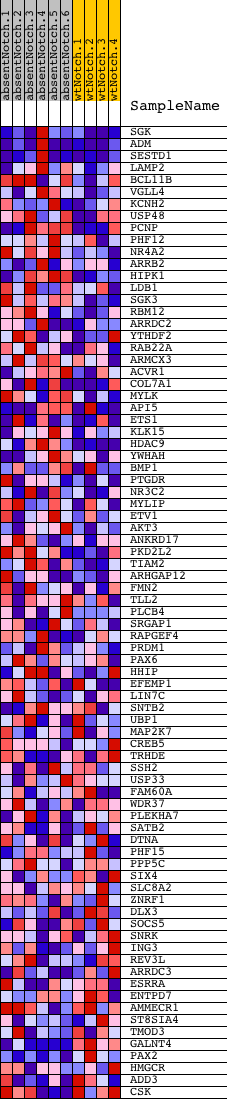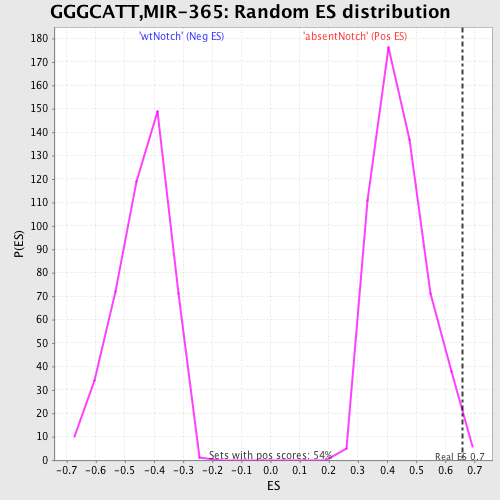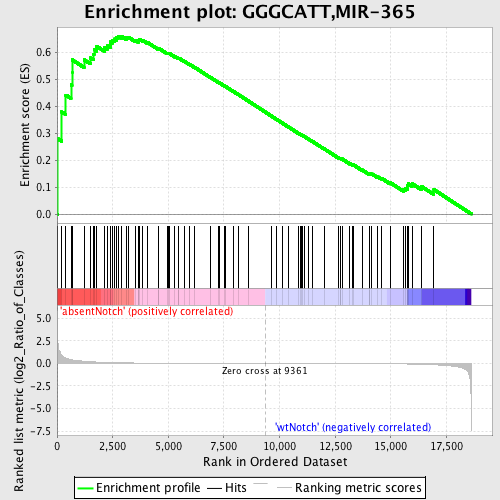
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | GGGCATT,MIR-365 |
| Enrichment Score (ES) | 0.660159 |
| Normalized Enrichment Score (NES) | 1.485502 |
| Nominal p-value | 0.009191177 |
| FDR q-value | 0.3488145 |
| FWER p-Value | 0.99 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SGK | 1400131 2480056 | 32 | 2.307 | 0.2791 | Yes | ||
| 2 | ADM | 4810121 | 206 | 0.898 | 0.3791 | Yes | ||
| 3 | SESTD1 | 1090333 3780593 | 391 | 0.596 | 0.4416 | Yes | ||
| 4 | LAMP2 | 1230402 1980373 | 626 | 0.413 | 0.4793 | Yes | ||
| 5 | BCL11B | 2680673 | 671 | 0.394 | 0.5249 | Yes | ||
| 6 | VGLL4 | 6860463 | 676 | 0.391 | 0.5723 | Yes | ||
| 7 | KCNH2 | 1170451 | 1214 | 0.239 | 0.5724 | Yes | ||
| 8 | USP48 | 2480010 | 1506 | 0.186 | 0.5793 | Yes | ||
| 9 | PCNP | 610768 | 1642 | 0.165 | 0.5921 | Yes | ||
| 10 | PHF12 | 870400 3130687 | 1663 | 0.162 | 0.6107 | Yes | ||
| 11 | NR4A2 | 60273 | 1781 | 0.148 | 0.6225 | Yes | ||
| 12 | ARRB2 | 2060441 | 2128 | 0.115 | 0.6178 | Yes | ||
| 13 | HIPK1 | 110193 | 2248 | 0.105 | 0.6242 | Yes | ||
| 14 | LDB1 | 5270601 | 2393 | 0.095 | 0.6280 | Yes | ||
| 15 | SGK3 | 1990408 4760452 5130278 6020594 6620471 | 2407 | 0.094 | 0.6387 | Yes | ||
| 16 | RBM12 | 4570239 6040411 6660129 | 2490 | 0.089 | 0.6451 | Yes | ||
| 17 | ARRDC2 | 520390 | 2594 | 0.082 | 0.6495 | Yes | ||
| 18 | YTHDF2 | 4060332 4610279 | 2678 | 0.077 | 0.6544 | Yes | ||
| 19 | RAB22A | 110500 3830707 | 2738 | 0.074 | 0.6602 | Yes | ||
| 20 | ARMCX3 | 3290309 | 2909 | 0.065 | 0.6589 | No | ||
| 21 | ACVR1 | 6840671 | 3108 | 0.056 | 0.6550 | No | ||
| 22 | COL7A1 | 1230168 2190072 | 3204 | 0.052 | 0.6562 | No | ||
| 23 | MYLK | 4010600 7000364 | 3505 | 0.042 | 0.6451 | No | ||
| 24 | API5 | 540593 1740114 6980692 | 3644 | 0.038 | 0.6423 | No | ||
| 25 | ETS1 | 5270278 6450717 6620465 | 3678 | 0.037 | 0.6451 | No | ||
| 26 | KLK15 | 6350086 | 3698 | 0.037 | 0.6486 | No | ||
| 27 | HDAC9 | 1990010 2260133 | 3838 | 0.034 | 0.6452 | No | ||
| 28 | YWHAH | 1660133 2810053 | 4048 | 0.030 | 0.6376 | No | ||
| 29 | BMP1 | 380594 2940576 3710593 | 4556 | 0.022 | 0.6129 | No | ||
| 30 | PTGDR | 3850161 | 4569 | 0.022 | 0.6150 | No | ||
| 31 | NR3C2 | 2480324 6400093 | 4961 | 0.018 | 0.5961 | No | ||
| 32 | MYLIP | 50717 4050672 | 4995 | 0.018 | 0.5965 | No | ||
| 33 | ETV1 | 70735 2940603 5080463 | 5034 | 0.017 | 0.5966 | No | ||
| 34 | AKT3 | 1580270 3290278 | 5297 | 0.015 | 0.5843 | No | ||
| 35 | ANKRD17 | 2120402 3850035 6400044 | 5433 | 0.014 | 0.5788 | No | ||
| 36 | PKD2L2 | 5270685 | 5472 | 0.014 | 0.5784 | No | ||
| 37 | TIAM2 | 360195 | 5703 | 0.013 | 0.5676 | No | ||
| 38 | ARHGAP12 | 1050181 | 5961 | 0.011 | 0.5551 | No | ||
| 39 | FMN2 | 450463 | 6170 | 0.010 | 0.5451 | No | ||
| 40 | TLL2 | 6220040 | 6913 | 0.007 | 0.5060 | No | ||
| 41 | PLCB4 | 50576 | 7253 | 0.006 | 0.4884 | No | ||
| 42 | SRGAP1 | 3940253 | 7317 | 0.006 | 0.4857 | No | ||
| 43 | RAPGEF4 | 6100563 | 7514 | 0.005 | 0.4757 | No | ||
| 44 | PRDM1 | 3170347 3520301 | 7566 | 0.005 | 0.4736 | No | ||
| 45 | PAX6 | 1190025 | 7927 | 0.004 | 0.4546 | No | ||
| 46 | HHIP | 6940438 | 8160 | 0.003 | 0.4425 | No | ||
| 47 | EFEMP1 | 5860324 | 8585 | 0.002 | 0.4199 | No | ||
| 48 | LIN7C | 4850341 | 9649 | -0.001 | 0.3626 | No | ||
| 49 | SNTB2 | 2630014 5390035 | 9842 | -0.001 | 0.3524 | No | ||
| 50 | UBP1 | 6020707 | 10139 | -0.002 | 0.3367 | No | ||
| 51 | MAP2K7 | 2260086 | 10416 | -0.003 | 0.3221 | No | ||
| 52 | CREB5 | 2320368 | 10828 | -0.004 | 0.3004 | No | ||
| 53 | TRHDE | 1690068 | 10932 | -0.004 | 0.2954 | No | ||
| 54 | SSH2 | 360056 | 10991 | -0.005 | 0.2928 | No | ||
| 55 | USP33 | 780047 | 11017 | -0.005 | 0.2920 | No | ||
| 56 | FAM60A | 3940092 | 11124 | -0.005 | 0.2869 | No | ||
| 57 | WDR37 | 2630519 6840347 | 11315 | -0.005 | 0.2773 | No | ||
| 58 | PLEKHA7 | 1410195 | 11481 | -0.006 | 0.2692 | No | ||
| 59 | SATB2 | 2470181 | 11996 | -0.008 | 0.2424 | No | ||
| 60 | DTNA | 1340600 1780731 2340278 2850132 | 12654 | -0.011 | 0.2083 | No | ||
| 61 | PHF15 | 6550066 6860673 | 12731 | -0.012 | 0.2056 | No | ||
| 62 | PPP5C | 3130047 | 12734 | -0.012 | 0.2069 | No | ||
| 63 | SIX4 | 6760022 | 12807 | -0.012 | 0.2045 | No | ||
| 64 | SLC8A2 | 6220092 | 13162 | -0.014 | 0.1871 | No | ||
| 65 | ZNRF1 | 70592 | 13281 | -0.015 | 0.1826 | No | ||
| 66 | DLX3 | 3190497 | 13310 | -0.016 | 0.1830 | No | ||
| 67 | SOCS5 | 3830398 7100093 | 13714 | -0.019 | 0.1636 | No | ||
| 68 | SNRK | 630021 2030731 6350017 | 14028 | -0.023 | 0.1495 | No | ||
| 69 | ING3 | 110438 2060390 | 14030 | -0.023 | 0.1522 | No | ||
| 70 | REV3L | 1090717 | 14151 | -0.024 | 0.1487 | No | ||
| 71 | ARRDC3 | 4610390 | 14417 | -0.028 | 0.1378 | No | ||
| 72 | ESRRA | 6550242 | 14569 | -0.031 | 0.1334 | No | ||
| 73 | ENTPD7 | 1980601 | 14978 | -0.040 | 0.1162 | No | ||
| 74 | AMMECR1 | 3800435 | 15575 | -0.059 | 0.0913 | No | ||
| 75 | ST8SIA4 | 2680605 3060215 | 15659 | -0.063 | 0.0944 | No | ||
| 76 | TMOD3 | 840088 | 15753 | -0.067 | 0.0975 | No | ||
| 77 | GALNT4 | 2970593 | 15756 | -0.067 | 0.1056 | No | ||
| 78 | PAX2 | 6040270 7000133 | 15775 | -0.068 | 0.1129 | No | ||
| 79 | HMGCR | 2630242 | 15955 | -0.077 | 0.1126 | No | ||
| 80 | ADD3 | 4150739 5390600 | 16385 | -0.104 | 0.1020 | No | ||
| 81 | CSK | 6350593 | 16935 | -0.150 | 0.0907 | No |