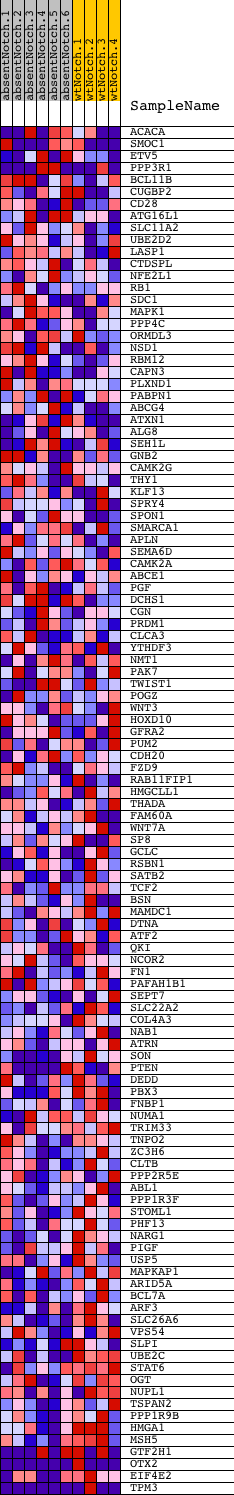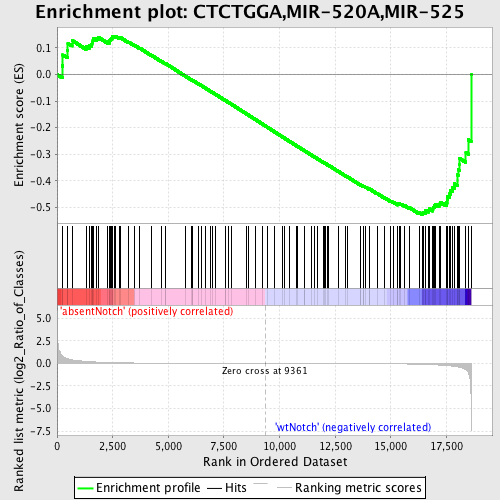
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | CTCTGGA,MIR-520A,MIR-525 |
| Enrichment Score (ES) | -0.526996 |
| Normalized Enrichment Score (NES) | -1.2870831 |
| Nominal p-value | 0.078431375 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ACACA | 2490612 2680369 | 235 | 0.843 | 0.0327 | No | ||
| 2 | SMOC1 | 840039 | 259 | 0.785 | 0.0737 | No | ||
| 3 | ETV5 | 110017 | 464 | 0.533 | 0.0913 | No | ||
| 4 | PPP3R1 | 1190041 1570487 | 487 | 0.504 | 0.1173 | No | ||
| 5 | BCL11B | 2680673 | 671 | 0.394 | 0.1287 | No | ||
| 6 | CUGBP2 | 6180121 6840139 | 1302 | 0.222 | 0.1066 | No | ||
| 7 | CD28 | 1400739 4210093 | 1442 | 0.195 | 0.1096 | No | ||
| 8 | ATG16L1 | 5390292 | 1543 | 0.179 | 0.1138 | No | ||
| 9 | SLC11A2 | 3140603 | 1569 | 0.175 | 0.1219 | No | ||
| 10 | UBE2D2 | 4590671 6510446 | 1611 | 0.169 | 0.1288 | No | ||
| 11 | LASP1 | 1690528 2190341 6420341 | 1635 | 0.166 | 0.1365 | No | ||
| 12 | CTDSPL | 4670546 | 1778 | 0.149 | 0.1368 | No | ||
| 13 | NFE2L1 | 6130053 | 1862 | 0.140 | 0.1399 | No | ||
| 14 | RB1 | 5900338 | 2280 | 0.103 | 0.1229 | No | ||
| 15 | SDC1 | 3440471 6350408 | 2364 | 0.097 | 0.1237 | No | ||
| 16 | MAPK1 | 3190193 6200253 | 2368 | 0.097 | 0.1287 | No | ||
| 17 | PPP4C | 5130593 | 2388 | 0.095 | 0.1328 | No | ||
| 18 | ORMDL3 | 1340711 | 2424 | 0.093 | 0.1359 | No | ||
| 19 | NSD1 | 4560519 4780672 | 2468 | 0.090 | 0.1384 | No | ||
| 20 | RBM12 | 4570239 6040411 6660129 | 2490 | 0.089 | 0.1421 | No | ||
| 21 | CAPN3 | 1980451 6940047 | 2569 | 0.083 | 0.1424 | No | ||
| 22 | PLXND1 | 5270682 | 2608 | 0.081 | 0.1447 | No | ||
| 23 | PABPN1 | 460519 | 2798 | 0.070 | 0.1382 | No | ||
| 24 | ABCG4 | 3440458 | 2854 | 0.067 | 0.1389 | No | ||
| 25 | ATXN1 | 5550156 | 3210 | 0.052 | 0.1225 | No | ||
| 26 | ALG8 | 4150497 4780500 6290563 | 3488 | 0.043 | 0.1098 | No | ||
| 27 | SEH1L | 4590563 | 3723 | 0.036 | 0.0991 | No | ||
| 28 | GNB2 | 2350053 | 4261 | 0.026 | 0.0715 | No | ||
| 29 | CAMK2G | 630097 | 4695 | 0.021 | 0.0492 | No | ||
| 30 | THY1 | 5910162 | 4874 | 0.019 | 0.0406 | No | ||
| 31 | KLF13 | 4670563 | 4876 | 0.019 | 0.0416 | No | ||
| 32 | SPRY4 | 1570594 | 5763 | 0.012 | -0.0057 | No | ||
| 33 | SPON1 | 1660301 | 6030 | 0.011 | -0.0194 | No | ||
| 34 | SMARCA1 | 4230315 | 6064 | 0.011 | -0.0207 | No | ||
| 35 | APLN | 6620632 | 6087 | 0.011 | -0.0213 | No | ||
| 36 | SEMA6D | 4050324 5860138 6350307 | 6351 | 0.009 | -0.0350 | No | ||
| 37 | CAMK2A | 1740333 1940112 4150292 | 6487 | 0.009 | -0.0418 | No | ||
| 38 | ABCE1 | 4280050 | 6682 | 0.008 | -0.0519 | No | ||
| 39 | PGF | 4810593 | 6896 | 0.007 | -0.0630 | No | ||
| 40 | DCHS1 | 4850487 | 6964 | 0.007 | -0.0662 | No | ||
| 41 | CGN | 5890139 6370167 6400537 | 7122 | 0.006 | -0.0744 | No | ||
| 42 | PRDM1 | 3170347 3520301 | 7566 | 0.005 | -0.0981 | No | ||
| 43 | CLCA3 | 6510167 | 7567 | 0.005 | -0.0978 | No | ||
| 44 | YTHDF3 | 4540632 4670717 7040609 | 7684 | 0.005 | -0.1038 | No | ||
| 45 | NMT1 | 5810152 | 7840 | 0.004 | -0.1120 | No | ||
| 46 | PAK7 | 3830164 | 8495 | 0.002 | -0.1472 | No | ||
| 47 | TWIST1 | 5130619 | 8586 | 0.002 | -0.1520 | No | ||
| 48 | POGZ | 870348 7040286 | 8907 | 0.001 | -0.1692 | No | ||
| 49 | WNT3 | 2360685 | 9219 | 0.000 | -0.1860 | No | ||
| 50 | HOXD10 | 6100039 | 9478 | -0.000 | -0.1999 | No | ||
| 51 | GFRA2 | 2350278 | 9776 | -0.001 | -0.2159 | No | ||
| 52 | PUM2 | 4200441 5910446 | 10115 | -0.002 | -0.2341 | No | ||
| 53 | CDH20 | 5420215 7100711 | 10227 | -0.002 | -0.2399 | No | ||
| 54 | FZD9 | 5360136 | 10464 | -0.003 | -0.2525 | No | ||
| 55 | RAB11FIP1 | 2470333 | 10751 | -0.004 | -0.2678 | No | ||
| 56 | HMGCLL1 | 2570091 | 10805 | -0.004 | -0.2704 | No | ||
| 57 | THADA | 6760348 | 11118 | -0.005 | -0.2870 | No | ||
| 58 | FAM60A | 3940092 | 11124 | -0.005 | -0.2870 | No | ||
| 59 | WNT7A | 1170315 | 11450 | -0.006 | -0.3043 | No | ||
| 60 | SP8 | 4060576 | 11585 | -0.007 | -0.3112 | No | ||
| 61 | GCLC | 2810731 | 11719 | -0.007 | -0.3180 | No | ||
| 62 | RSBN1 | 7000487 | 11968 | -0.008 | -0.3310 | No | ||
| 63 | SATB2 | 2470181 | 11996 | -0.008 | -0.3320 | No | ||
| 64 | TCF2 | 870338 5050632 | 12076 | -0.008 | -0.3358 | No | ||
| 65 | BSN | 5270347 | 12175 | -0.009 | -0.3406 | No | ||
| 66 | MAMDC1 | 3710129 4480075 | 12209 | -0.009 | -0.3419 | No | ||
| 67 | DTNA | 1340600 1780731 2340278 2850132 | 12654 | -0.011 | -0.3653 | No | ||
| 68 | ATF2 | 2260348 2480441 | 12975 | -0.013 | -0.3819 | No | ||
| 69 | QKI | 5220093 6130044 | 13044 | -0.014 | -0.3849 | No | ||
| 70 | NCOR2 | 1980575 6550050 | 13627 | -0.018 | -0.4153 | No | ||
| 71 | FN1 | 1170601 2970647 6220288 6940037 | 13644 | -0.018 | -0.4152 | No | ||
| 72 | PAFAH1B1 | 4230333 6420121 6450066 | 13768 | -0.020 | -0.4208 | No | ||
| 73 | SEPT7 | 2760685 | 13772 | -0.020 | -0.4199 | No | ||
| 74 | SLC22A2 | 5670520 | 13871 | -0.021 | -0.4241 | No | ||
| 75 | COL4A3 | 5910075 | 14021 | -0.023 | -0.4309 | No | ||
| 76 | NAB1 | 430739 1230114 3390736 6660056 | 14042 | -0.023 | -0.4308 | No | ||
| 77 | ATRN | 870735 3360390 | 14385 | -0.028 | -0.4478 | No | ||
| 78 | SON | 3120148 5860239 7040403 | 14716 | -0.034 | -0.4638 | No | ||
| 79 | PTEN | 3390064 | 15006 | -0.040 | -0.4772 | No | ||
| 80 | DEDD | 6400131 | 15126 | -0.044 | -0.4813 | No | ||
| 81 | PBX3 | 1300424 3710577 6180575 | 15313 | -0.050 | -0.4887 | No | ||
| 82 | FNBP1 | 6100017 | 15314 | -0.050 | -0.4860 | No | ||
| 83 | NUMA1 | 6520576 | 15378 | -0.052 | -0.4866 | No | ||
| 84 | TRIM33 | 580619 2230280 3990433 6200747 | 15451 | -0.055 | -0.4875 | No | ||
| 85 | TNPO2 | 3840164 | 15611 | -0.061 | -0.4929 | No | ||
| 86 | ZC3H6 | 130711 | 15840 | -0.071 | -0.5014 | No | ||
| 87 | CLTB | 5550037 | 16279 | -0.096 | -0.5199 | No | ||
| 88 | PPP2R5E | 70671 2120056 | 16412 | -0.106 | -0.5213 | Yes | ||
| 89 | ABL1 | 1050593 2030050 4010114 | 16464 | -0.110 | -0.5181 | Yes | ||
| 90 | PPP1R3F | 2450114 | 16550 | -0.116 | -0.5165 | Yes | ||
| 91 | STOML1 | 870452 5360131 | 16570 | -0.117 | -0.5112 | Yes | ||
| 92 | PHF13 | 450435 | 16695 | -0.127 | -0.5110 | Yes | ||
| 93 | NARG1 | 5910563 6350095 | 16732 | -0.131 | -0.5060 | Yes | ||
| 94 | PIGF | 5690050 | 16882 | -0.144 | -0.5063 | Yes | ||
| 95 | USP5 | 4850017 | 16896 | -0.145 | -0.4991 | Yes | ||
| 96 | MAPKAP1 | 1660161 2450575 | 16953 | -0.152 | -0.4940 | Yes | ||
| 97 | ARID5A | 2450133 | 17007 | -0.158 | -0.4883 | Yes | ||
| 98 | BCL7A | 4560095 | 17170 | -0.178 | -0.4875 | Yes | ||
| 99 | ARF3 | 3710446 | 17219 | -0.187 | -0.4800 | Yes | ||
| 100 | SLC26A6 | 4810575 | 17492 | -0.236 | -0.4820 | Yes | ||
| 101 | VPS54 | 1660168 | 17527 | -0.242 | -0.4707 | Yes | ||
| 102 | SLPI | 2120446 | 17553 | -0.248 | -0.4588 | Yes | ||
| 103 | UBE2C | 6130017 | 17653 | -0.271 | -0.4495 | Yes | ||
| 104 | STAT6 | 1190010 5720019 | 17670 | -0.275 | -0.4356 | Yes | ||
| 105 | OGT | 2360131 4610333 | 17770 | -0.304 | -0.4246 | Yes | ||
| 106 | NUPL1 | 360273 6770100 | 17855 | -0.331 | -0.4113 | Yes | ||
| 107 | TSPAN2 | 3940161 | 18007 | -0.386 | -0.3987 | Yes | ||
| 108 | PPP1R9B | 3130619 | 18008 | -0.386 | -0.3779 | Yes | ||
| 109 | HMGA1 | 6580408 | 18028 | -0.394 | -0.3577 | Yes | ||
| 110 | MSH5 | 1340019 4730333 | 18092 | -0.417 | -0.3386 | Yes | ||
| 111 | GTF2H1 | 2570746 2650148 | 18098 | -0.422 | -0.3162 | Yes | ||
| 112 | OTX2 | 1190471 | 18377 | -0.689 | -0.2941 | Yes | ||
| 113 | EIF4E2 | 2760072 5910609 | 18487 | -1.013 | -0.2454 | Yes | ||
| 114 | TPM3 | 670600 3170296 5670167 6650471 | 18609 | -4.686 | 0.0004 | Yes |