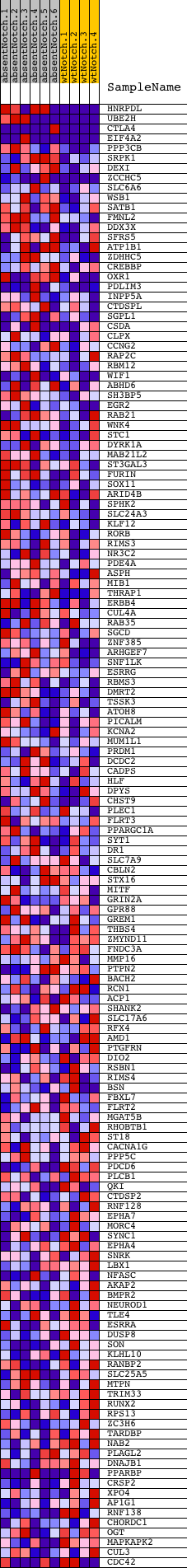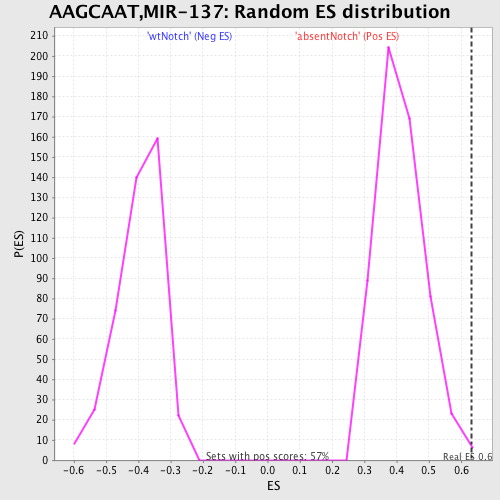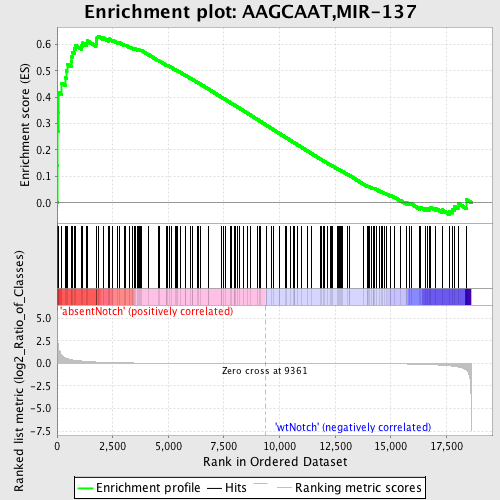
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | AAGCAAT,MIR-137 |
| Enrichment Score (ES) | 0.63115513 |
| Normalized Enrichment Score (NES) | 1.5359608 |
| Nominal p-value | 0.0017482517 |
| FDR q-value | 0.26650125 |
| FWER p-Value | 0.913 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HNRPDL | 1050102 1090181 5360471 | 19 | 2.927 | 0.1414 | Yes | ||
| 2 | UBE2H | 1980142 2970079 | 24 | 2.659 | 0.2706 | Yes | ||
| 3 | CTLA4 | 6590537 | 69 | 1.537 | 0.3430 | Yes | ||
| 4 | EIF4A2 | 1170494 1740711 2850504 | 72 | 1.519 | 0.4169 | Yes | ||
| 5 | PPP3CB | 6020156 | 211 | 0.891 | 0.4528 | Yes | ||
| 6 | SRPK1 | 450110 | 382 | 0.611 | 0.4733 | Yes | ||
| 7 | DEXI | 6220358 6550685 | 415 | 0.575 | 0.4996 | Yes | ||
| 8 | ZCCHC5 | 2650021 | 445 | 0.549 | 0.5247 | Yes | ||
| 9 | SLC6A6 | 6100300 | 635 | 0.409 | 0.5344 | Yes | ||
| 10 | WSB1 | 870563 | 661 | 0.399 | 0.5524 | Yes | ||
| 11 | SATB1 | 5670154 | 678 | 0.390 | 0.5706 | Yes | ||
| 12 | FMNL2 | 4060438 | 770 | 0.354 | 0.5829 | Yes | ||
| 13 | DDX3X | 2190020 | 842 | 0.330 | 0.5951 | Yes | ||
| 14 | SFRS5 | 3450176 6350008 | 1094 | 0.262 | 0.5942 | Yes | ||
| 15 | ATP1B1 | 3130594 | 1123 | 0.256 | 0.6052 | Yes | ||
| 16 | ZDHHC5 | 1690711 | 1330 | 0.218 | 0.6047 | Yes | ||
| 17 | CREBBP | 5690035 7040050 | 1349 | 0.214 | 0.6141 | Yes | ||
| 18 | OXR1 | 2370039 3190706 | 1748 | 0.153 | 0.6000 | Yes | ||
| 19 | PDLIM3 | 1410095 | 1754 | 0.152 | 0.6071 | Yes | ||
| 20 | INPP5A | 5910195 | 1772 | 0.150 | 0.6135 | Yes | ||
| 21 | CTDSPL | 4670546 | 1778 | 0.149 | 0.6205 | Yes | ||
| 22 | SGPL1 | 4480059 | 1791 | 0.147 | 0.6270 | Yes | ||
| 23 | CSDA | 2470092 3060053 4780056 | 1843 | 0.142 | 0.6312 | Yes | ||
| 24 | CLPX | 6220156 | 2062 | 0.120 | 0.6252 | No | ||
| 25 | CCNG2 | 3190095 | 2305 | 0.102 | 0.6171 | No | ||
| 26 | RAP2C | 1690132 | 2333 | 0.099 | 0.6204 | No | ||
| 27 | RBM12 | 4570239 6040411 6660129 | 2490 | 0.089 | 0.6163 | No | ||
| 28 | WIF1 | 7100184 | 2734 | 0.074 | 0.6067 | No | ||
| 29 | ABHD6 | 630717 | 2809 | 0.069 | 0.6061 | No | ||
| 30 | SH3BP5 | 7100408 | 3022 | 0.060 | 0.5975 | No | ||
| 31 | EGR2 | 3800403 | 3085 | 0.057 | 0.5969 | No | ||
| 32 | RAB21 | 5570070 | 3272 | 0.049 | 0.5893 | No | ||
| 33 | WNK4 | 1240682 | 3378 | 0.046 | 0.5858 | No | ||
| 34 | STC1 | 360161 | 3492 | 0.042 | 0.5817 | No | ||
| 35 | DYRK1A | 3190181 | 3512 | 0.042 | 0.5828 | No | ||
| 36 | MAB21L2 | 1770605 | 3597 | 0.039 | 0.5801 | No | ||
| 37 | ST3GAL3 | 2810601 | 3658 | 0.038 | 0.5787 | No | ||
| 38 | FURIN | 4120168 | 3692 | 0.037 | 0.5787 | No | ||
| 39 | SOX11 | 610279 | 3746 | 0.036 | 0.5776 | No | ||
| 40 | ARID4B | 730754 6660026 | 3781 | 0.035 | 0.5775 | No | ||
| 41 | SPHK2 | 2030487 4120670 4850114 | 4121 | 0.029 | 0.5605 | No | ||
| 42 | SLC24A3 | 5080707 | 4537 | 0.023 | 0.5392 | No | ||
| 43 | KLF12 | 1660095 4810288 5340546 6520286 | 4618 | 0.022 | 0.5359 | No | ||
| 44 | RORB | 6400035 | 4913 | 0.018 | 0.5209 | No | ||
| 45 | RIMS3 | 3890563 | 4933 | 0.018 | 0.5207 | No | ||
| 46 | NR3C2 | 2480324 6400093 | 4961 | 0.018 | 0.5202 | No | ||
| 47 | PDE4A | 1190619 1340129 5720100 6370035 | 5036 | 0.017 | 0.5170 | No | ||
| 48 | ASPH | 360722 1940039 2100066 2340121 2650113 2810064 2970008 3140040 3800750 4210014 4280133 4590035 4760433 6370671 6940338 | 5131 | 0.017 | 0.5127 | No | ||
| 49 | MIB1 | 3800537 | 5336 | 0.015 | 0.5024 | No | ||
| 50 | THRAP1 | 2470121 | 5348 | 0.015 | 0.5025 | No | ||
| 51 | ERBB4 | 4610400 4810017 | 5389 | 0.015 | 0.5011 | No | ||
| 52 | CUL4A | 1170088 2320008 2470278 | 5523 | 0.014 | 0.4946 | No | ||
| 53 | RAB35 | 630056 | 5761 | 0.012 | 0.4823 | No | ||
| 54 | SGCD | 2940685 5080753 | 5777 | 0.012 | 0.4821 | No | ||
| 55 | ZNF385 | 450176 4810358 | 5992 | 0.011 | 0.4711 | No | ||
| 56 | ARHGEF7 | 1690441 | 6095 | 0.011 | 0.4661 | No | ||
| 57 | SNF1LK | 6110403 | 6102 | 0.011 | 0.4663 | No | ||
| 58 | ESRRG | 4010181 | 6290 | 0.010 | 0.4566 | No | ||
| 59 | RBMS3 | 6110722 | 6355 | 0.009 | 0.4536 | No | ||
| 60 | DMRT2 | 2630068 | 6461 | 0.009 | 0.4483 | No | ||
| 61 | TSSK3 | 1740673 | 6784 | 0.008 | 0.4313 | No | ||
| 62 | ATOH8 | 4540204 | 6787 | 0.008 | 0.4315 | No | ||
| 63 | PICALM | 1940280 | 7377 | 0.005 | 0.3999 | No | ||
| 64 | KCNA2 | 5080403 | 7391 | 0.005 | 0.3995 | No | ||
| 65 | MUM1L1 | 4010411 | 7472 | 0.005 | 0.3954 | No | ||
| 66 | PRDM1 | 3170347 3520301 | 7566 | 0.005 | 0.3906 | No | ||
| 67 | DCDC2 | 7000338 | 7805 | 0.004 | 0.3779 | No | ||
| 68 | CADPS | 2320181 | 7837 | 0.004 | 0.3764 | No | ||
| 69 | HLF | 2370113 | 7951 | 0.004 | 0.3705 | No | ||
| 70 | DPYS | 3130670 | 7985 | 0.004 | 0.3689 | No | ||
| 71 | CHST9 | 4560092 | 8002 | 0.004 | 0.3682 | No | ||
| 72 | PLEC1 | 6520292 50500 380075 1570088 2510692 2630497 2680017 2900746 4560497 4850376 6290398 7040471 | 8100 | 0.003 | 0.3631 | No | ||
| 73 | FLRT3 | 70685 2760497 6040519 | 8214 | 0.003 | 0.3572 | No | ||
| 74 | PPARGC1A | 4670040 | 8390 | 0.003 | 0.3478 | No | ||
| 75 | SYT1 | 840364 | 8535 | 0.002 | 0.3401 | No | ||
| 76 | DR1 | 5860750 | 8694 | 0.002 | 0.3316 | No | ||
| 77 | SLC7A9 | 5420735 5700450 | 8992 | 0.001 | 0.3156 | No | ||
| 78 | CBLN2 | 4760132 | 9111 | 0.001 | 0.3093 | No | ||
| 79 | STX16 | 70315 | 9143 | 0.001 | 0.3076 | No | ||
| 80 | MITF | 380056 | 9394 | -0.000 | 0.2941 | No | ||
| 81 | GRIN2A | 6550538 | 9404 | -0.000 | 0.2936 | No | ||
| 82 | GPR88 | 2450435 | 9639 | -0.001 | 0.2810 | No | ||
| 83 | GREM1 | 3940180 | 9716 | -0.001 | 0.2769 | No | ||
| 84 | THBS4 | 3710348 | 10001 | -0.002 | 0.2616 | No | ||
| 85 | ZMYND11 | 630181 2570019 4850138 5360040 | 10007 | -0.002 | 0.2614 | No | ||
| 86 | FNDC3A | 2690097 3290044 | 10265 | -0.002 | 0.2476 | No | ||
| 87 | MMP16 | 2680139 | 10299 | -0.002 | 0.2459 | No | ||
| 88 | PTPN2 | 130315 | 10496 | -0.003 | 0.2355 | No | ||
| 89 | BACH2 | 6760131 | 10634 | -0.003 | 0.2282 | No | ||
| 90 | RCN1 | 2480041 | 10657 | -0.003 | 0.2272 | No | ||
| 91 | ACP1 | 1990435 | 10675 | -0.004 | 0.2264 | No | ||
| 92 | SHANK2 | 1500452 | 10822 | -0.004 | 0.2187 | No | ||
| 93 | SLC17A6 | 4210576 | 10970 | -0.004 | 0.2110 | No | ||
| 94 | RFX4 | 3710100 5340215 | 11260 | -0.005 | 0.1956 | No | ||
| 95 | AMD1 | 6290128 | 11451 | -0.006 | 0.1856 | No | ||
| 96 | PTGFRN | 4120524 | 11831 | -0.007 | 0.1654 | No | ||
| 97 | DIO2 | 380504 540403 | 11896 | -0.008 | 0.1623 | No | ||
| 98 | RSBN1 | 7000487 | 11968 | -0.008 | 0.1589 | No | ||
| 99 | RIMS4 | 6110500 | 12025 | -0.008 | 0.1563 | No | ||
| 100 | BSN | 5270347 | 12175 | -0.009 | 0.1486 | No | ||
| 101 | FBXL7 | 6620711 | 12306 | -0.009 | 0.1420 | No | ||
| 102 | FLRT2 | 4200035 | 12343 | -0.010 | 0.1405 | No | ||
| 103 | MGAT5B | 380131 2570364 | 12379 | -0.010 | 0.1391 | No | ||
| 104 | RHOBTB1 | 1410010 | 12607 | -0.011 | 0.1274 | No | ||
| 105 | ST18 | 5720537 | 12653 | -0.011 | 0.1255 | No | ||
| 106 | CACNA1G | 3060161 4810706 5550722 | 12693 | -0.011 | 0.1239 | No | ||
| 107 | PPP5C | 3130047 | 12734 | -0.012 | 0.1223 | No | ||
| 108 | PDCD6 | 4760239 | 12779 | -0.012 | 0.1205 | No | ||
| 109 | PLCB1 | 2570050 7100102 | 12828 | -0.012 | 0.1185 | No | ||
| 110 | QKI | 5220093 6130044 | 13044 | -0.014 | 0.1075 | No | ||
| 111 | CTDSP2 | 5340315 5690133 | 13127 | -0.014 | 0.1038 | No | ||
| 112 | RNF128 | 1230300 5670273 | 13141 | -0.014 | 0.1037 | No | ||
| 113 | EPHA7 | 1190288 4010278 | 13765 | -0.020 | 0.0710 | No | ||
| 114 | MORC4 | 1740040 | 13937 | -0.022 | 0.0628 | No | ||
| 115 | SYNC1 | 580114 | 13946 | -0.022 | 0.0634 | No | ||
| 116 | EPHA4 | 460750 | 14002 | -0.022 | 0.0615 | No | ||
| 117 | SNRK | 630021 2030731 6350017 | 14028 | -0.023 | 0.0612 | No | ||
| 118 | LBX1 | 1300164 4060204 | 14140 | -0.024 | 0.0564 | No | ||
| 119 | NFASC | 1940619 2470156 4590168 6840524 | 14208 | -0.025 | 0.0540 | No | ||
| 120 | AKAP2 | 5550347 | 14230 | -0.025 | 0.0541 | No | ||
| 121 | BMPR2 | 5340066 6960328 | 14260 | -0.026 | 0.0538 | No | ||
| 122 | NEUROD1 | 3060619 | 14357 | -0.027 | 0.0499 | No | ||
| 123 | TLE4 | 1450064 1500519 | 14481 | -0.029 | 0.0447 | No | ||
| 124 | ESRRA | 6550242 | 14569 | -0.031 | 0.0415 | No | ||
| 125 | DUSP8 | 6650039 | 14633 | -0.032 | 0.0396 | No | ||
| 126 | SON | 3120148 5860239 7040403 | 14716 | -0.034 | 0.0368 | No | ||
| 127 | KLHL10 | 780139 | 14827 | -0.036 | 0.0326 | No | ||
| 128 | RANBP2 | 4280338 | 14964 | -0.039 | 0.0271 | No | ||
| 129 | SLC25A5 | 1980672 | 14983 | -0.040 | 0.0281 | No | ||
| 130 | MTPN | 3940167 | 15160 | -0.045 | 0.0207 | No | ||
| 131 | TRIM33 | 580619 2230280 3990433 6200747 | 15451 | -0.055 | 0.0077 | No | ||
| 132 | RUNX2 | 1400193 4230215 | 15688 | -0.064 | -0.0020 | No | ||
| 133 | RPS13 | 1570364 | 15694 | -0.064 | 0.0009 | No | ||
| 134 | ZC3H6 | 130711 | 15840 | -0.071 | -0.0035 | No | ||
| 135 | TARDBP | 870390 2350093 | 15911 | -0.075 | -0.0037 | No | ||
| 136 | NAB2 | 1570014 | 16292 | -0.097 | -0.0196 | No | ||
| 137 | PLAGL2 | 1770148 | 16329 | -0.099 | -0.0167 | No | ||
| 138 | DNAJB1 | 1090041 | 16558 | -0.117 | -0.0233 | No | ||
| 139 | PPARBP | 4280131 5690022 5860050 | 16642 | -0.122 | -0.0219 | No | ||
| 140 | CRSP2 | 1940059 | 16752 | -0.132 | -0.0213 | No | ||
| 141 | XPO4 | 3840551 | 16793 | -0.136 | -0.0169 | No | ||
| 142 | AP1G1 | 2360671 | 17023 | -0.159 | -0.0215 | No | ||
| 143 | RNF138 | 110142 3870129 6020441 6400193 | 17309 | -0.204 | -0.0270 | No | ||
| 144 | CHORDC1 | 6650577 | 17634 | -0.266 | -0.0316 | No | ||
| 145 | OGT | 2360131 4610333 | 17770 | -0.304 | -0.0241 | No | ||
| 146 | MAPKAPK2 | 2850500 | 17873 | -0.336 | -0.0133 | No | ||
| 147 | CUL3 | 1850520 | 18023 | -0.392 | -0.0023 | No | ||
| 148 | CDC42 | 1240168 3440278 4480519 5290162 | 18387 | -0.705 | 0.0124 | No |