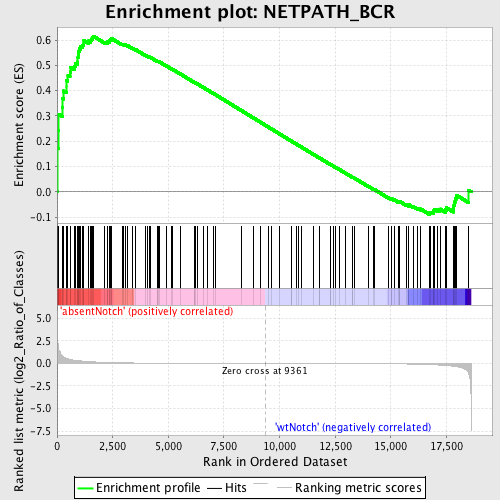
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | NETPATH_BCR |
| Enrichment Score (ES) | 0.61542386 |
| Normalized Enrichment Score (NES) | 1.4584616 |
| Nominal p-value | 0.010291595 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GTF2I | 2470750 3870408 3830369 3990082 4610138 | 7 | 3.986 | 0.1741 | Yes | ||
| 2 | CCND2 | 5340167 | 59 | 1.612 | 0.2419 | Yes | ||
| 3 | CDK2 | 130484 2260301 4010088 5050110 | 75 | 1.505 | 0.3069 | Yes | ||
| 4 | FCGR2B | 780750 | 245 | 0.825 | 0.3338 | Yes | ||
| 5 | PTK2 | 1780148 | 252 | 0.808 | 0.3689 | Yes | ||
| 6 | BCL2L11 | 780044 4200601 | 293 | 0.733 | 0.3988 | Yes | ||
| 7 | ITK | 2230592 | 434 | 0.562 | 0.4158 | Yes | ||
| 8 | CCNE1 | 110064 | 435 | 0.561 | 0.4403 | Yes | ||
| 9 | PPP3R1 | 1190041 1570487 | 487 | 0.504 | 0.4596 | Yes | ||
| 10 | SH3BP2 | 610131 | 581 | 0.438 | 0.4738 | Yes | ||
| 11 | CCND3 | 380528 730500 | 590 | 0.433 | 0.4923 | Yes | ||
| 12 | NFKBIA | 1570152 | 786 | 0.347 | 0.4969 | Yes | ||
| 13 | CDK4 | 540075 4540600 | 832 | 0.332 | 0.5090 | Yes | ||
| 14 | LAT2 | 5340440 | 931 | 0.304 | 0.5170 | Yes | ||
| 15 | NFATC3 | 5050520 | 935 | 0.303 | 0.5301 | Yes | ||
| 16 | CD72 | 2340369 | 945 | 0.297 | 0.5426 | Yes | ||
| 17 | PDPK1 | 6650168 | 961 | 0.293 | 0.5546 | Yes | ||
| 18 | CASP7 | 4210156 | 993 | 0.285 | 0.5654 | Yes | ||
| 19 | MAPK8 | 2640195 | 1063 | 0.270 | 0.5735 | Yes | ||
| 20 | PIP5K1C | 5890164 6510148 | 1148 | 0.251 | 0.5799 | Yes | ||
| 21 | CBL | 6380068 | 1202 | 0.241 | 0.5876 | Yes | ||
| 22 | BCL6 | 940100 | 1204 | 0.240 | 0.5981 | Yes | ||
| 23 | GSK3B | 5360348 | 1401 | 0.203 | 0.5964 | Yes | ||
| 24 | BAX | 3830008 | 1486 | 0.188 | 0.6001 | Yes | ||
| 25 | GRB2 | 6650398 | 1555 | 0.177 | 0.6041 | Yes | ||
| 26 | BTK | 3130044 | 1576 | 0.175 | 0.6107 | Yes | ||
| 27 | NEDD9 | 1740373 4230053 | 1625 | 0.167 | 0.6154 | Yes | ||
| 28 | CDK6 | 4920253 | 2151 | 0.113 | 0.5920 | No | ||
| 29 | CHUK | 7050736 | 2267 | 0.104 | 0.5903 | No | ||
| 30 | RB1 | 5900338 | 2280 | 0.103 | 0.5942 | No | ||
| 31 | CD79B | 1450066 3390358 | 2352 | 0.098 | 0.5946 | No | ||
| 32 | MAPK1 | 3190193 6200253 | 2368 | 0.097 | 0.5981 | No | ||
| 33 | GAB1 | 2970156 | 2398 | 0.094 | 0.6006 | No | ||
| 34 | MAPK14 | 5290731 | 2442 | 0.092 | 0.6023 | No | ||
| 35 | PTPRC | 130402 5290148 | 2453 | 0.091 | 0.6058 | No | ||
| 36 | CRKL | 4050427 | 2922 | 0.064 | 0.5833 | No | ||
| 37 | PTK2B | 4730411 | 2982 | 0.062 | 0.5828 | No | ||
| 38 | FYN | 2100468 4760520 4850687 | 3063 | 0.058 | 0.5810 | No | ||
| 39 | RAPGEF1 | 60040 4590670 | 3185 | 0.053 | 0.5768 | No | ||
| 40 | CD79A | 3450563 | 3401 | 0.045 | 0.5671 | No | ||
| 41 | BLNK | 70288 6590092 | 3535 | 0.041 | 0.5618 | No | ||
| 42 | JUN | 840170 | 3990 | 0.031 | 0.5386 | No | ||
| 43 | PLCG1 | 6020369 | 4062 | 0.030 | 0.5360 | No | ||
| 44 | CD22 | 3120239 | 4139 | 0.028 | 0.5332 | No | ||
| 45 | BCL2 | 730132 1570736 2470138 3800044 4810037 5690068 5860504 6650164 | 4210 | 0.027 | 0.5306 | No | ||
| 46 | CD81 | 5270093 | 4509 | 0.023 | 0.5155 | No | ||
| 47 | PRKD1 | 1410706 5090438 | 4512 | 0.023 | 0.5164 | No | ||
| 48 | PRKCD | 770592 | 4555 | 0.022 | 0.5151 | No | ||
| 49 | DOK3 | 130347 3870167 | 4609 | 0.022 | 0.5132 | No | ||
| 50 | MAP3K7 | 6040068 | 4923 | 0.018 | 0.4971 | No | ||
| 51 | PIK3AP1 | 6760008 | 5159 | 0.016 | 0.4851 | No | ||
| 52 | RPS6KB1 | 1450427 5080110 6200563 | 5175 | 0.016 | 0.4850 | No | ||
| 53 | CARD11 | 70338 | 5527 | 0.014 | 0.4666 | No | ||
| 54 | RASGRP3 | 6020504 | 6154 | 0.010 | 0.4332 | No | ||
| 55 | PRKCE | 5700053 | 6191 | 0.010 | 0.4317 | No | ||
| 56 | SYK | 6940133 | 6233 | 0.010 | 0.4299 | No | ||
| 57 | CD19 | 6940692 | 6315 | 0.010 | 0.4260 | No | ||
| 58 | BLK | 1940128 5390053 | 6594 | 0.008 | 0.4113 | No | ||
| 59 | BCAR1 | 1340215 | 6775 | 0.008 | 0.4019 | No | ||
| 60 | SHC1 | 2900731 3170504 6520537 | 7016 | 0.007 | 0.3892 | No | ||
| 61 | PTPN11 | 2230100 2470180 6100528 | 7100 | 0.006 | 0.3850 | No | ||
| 62 | HCK | 4230592 | 8298 | 0.003 | 0.3205 | No | ||
| 63 | ACTR3 | 1400497 | 8814 | 0.001 | 0.2927 | No | ||
| 64 | CDK7 | 2640451 | 9152 | 0.001 | 0.2745 | No | ||
| 65 | AKT1 | 5290746 | 9501 | -0.000 | 0.2557 | No | ||
| 66 | CR2 | 2850332 6450397 | 9615 | -0.001 | 0.2496 | No | ||
| 67 | PIK3R1 | 4730671 | 9982 | -0.002 | 0.2299 | No | ||
| 68 | GAB2 | 1410280 2340520 4280040 | 10514 | -0.003 | 0.2014 | No | ||
| 69 | CRK | 1230162 4780128 | 10527 | -0.003 | 0.2008 | No | ||
| 70 | PRKCQ | 2260170 3870193 | 10743 | -0.004 | 0.1894 | No | ||
| 71 | LCP2 | 2680066 6650707 | 10865 | -0.004 | 0.1830 | No | ||
| 72 | MAPK4 | 870142 | 10984 | -0.005 | 0.1768 | No | ||
| 73 | DOK1 | 1050279 2680112 5220273 | 11529 | -0.006 | 0.1477 | No | ||
| 74 | TEC | 1400576 | 11773 | -0.007 | 0.1349 | No | ||
| 75 | ITPR2 | 5360128 | 11774 | -0.007 | 0.1352 | No | ||
| 76 | CASP9 | 60577 1170368 2060576 | 12289 | -0.009 | 0.1078 | No | ||
| 77 | NCK1 | 6200575 6510050 | 12298 | -0.009 | 0.1078 | No | ||
| 78 | PDK2 | 2690017 | 12429 | -0.010 | 0.1012 | No | ||
| 79 | LYN | 6040600 | 12529 | -0.010 | 0.0963 | No | ||
| 80 | PIK3R2 | 1660193 | 12709 | -0.011 | 0.0872 | No | ||
| 81 | ATF2 | 2260348 2480441 | 12975 | -0.013 | 0.0734 | No | ||
| 82 | ACTR2 | 1170332 | 13265 | -0.015 | 0.0585 | No | ||
| 83 | PTPN18 | 2640537 | 13288 | -0.015 | 0.0579 | No | ||
| 84 | CREB1 | 1500717 2230358 3610600 6550601 | 13388 | -0.016 | 0.0533 | No | ||
| 85 | NFATC2 | 70450 540097 | 13983 | -0.022 | 0.0222 | No | ||
| 86 | BCL10 | 2360397 | 14211 | -0.025 | 0.0110 | No | ||
| 87 | IKBKG | 3450092 3840377 6590592 | 14249 | -0.026 | 0.0101 | No | ||
| 88 | SOS1 | 7050338 | 14900 | -0.037 | -0.0234 | No | ||
| 89 | PPP3CA | 4760332 6760092 | 15017 | -0.041 | -0.0279 | No | ||
| 90 | DAPP1 | 2480086 2970300 4480494 | 15020 | -0.041 | -0.0262 | No | ||
| 91 | CCNA2 | 5290075 | 15143 | -0.044 | -0.0309 | No | ||
| 92 | CBLB | 3520465 4670348 | 15328 | -0.050 | -0.0386 | No | ||
| 93 | RPS6KA1 | 4120647 | 15400 | -0.053 | -0.0401 | No | ||
| 94 | MAP4K1 | 4060692 | 15404 | -0.053 | -0.0380 | No | ||
| 95 | CYCS | 2030072 | 15708 | -0.064 | -0.0515 | No | ||
| 96 | LCK | 3360142 | 15797 | -0.069 | -0.0533 | No | ||
| 97 | REL | 360707 | 15811 | -0.069 | -0.0510 | No | ||
| 98 | VAV2 | 3610725 5890717 | 15998 | -0.079 | -0.0575 | No | ||
| 99 | NFATC1 | 510400 2320348 4050600 6180161 6290136 6620086 | 16205 | -0.091 | -0.0647 | No | ||
| 100 | IKBKB | 6840072 | 16337 | -0.100 | -0.0674 | No | ||
| 101 | RELA | 3830075 | 16742 | -0.132 | -0.0835 | No | ||
| 102 | VAV1 | 6020487 | 16797 | -0.137 | -0.0804 | No | ||
| 103 | GSK3A | 3140735 | 16905 | -0.147 | -0.0798 | No | ||
| 104 | CSK | 6350593 | 16935 | -0.150 | -0.0748 | No | ||
| 105 | HDAC5 | 2370035 6370139 | 16947 | -0.151 | -0.0688 | No | ||
| 106 | PTPN6 | 4230128 | 17084 | -0.167 | -0.0688 | No | ||
| 107 | SOS2 | 2060722 | 17212 | -0.185 | -0.0675 | No | ||
| 108 | RPS6 | 2060168 | 17457 | -0.230 | -0.0707 | No | ||
| 109 | ZAP70 | 1410494 2260504 | 17495 | -0.237 | -0.0623 | No | ||
| 110 | MAPK3 | 580161 4780035 | 17816 | -0.318 | -0.0657 | No | ||
| 111 | HCLS1 | 7100309 | 17834 | -0.324 | -0.0524 | No | ||
| 112 | MAPKAPK2 | 2850500 | 17873 | -0.336 | -0.0398 | No | ||
| 113 | WAS | 5270193 | 17888 | -0.343 | -0.0255 | No | ||
| 114 | RASA1 | 1240315 | 17940 | -0.363 | -0.0124 | No | ||
| 115 | CD5 | 6590338 | 18504 | -1.117 | 0.0061 | No |

