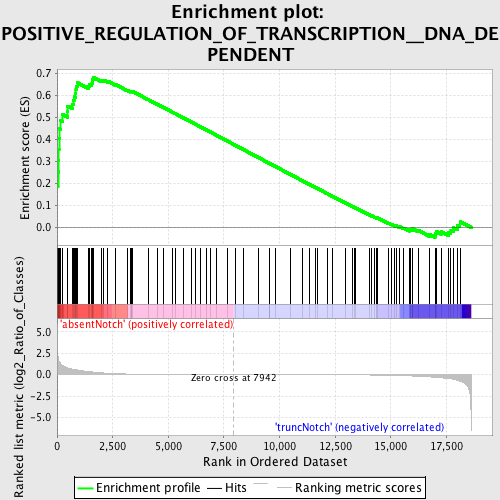
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | POSITIVE_REGULATION_OF_TRANSCRIPTION__DNA_DEPENDENT |
| Enrichment Score (ES) | 0.6821518 |
| Normalized Enrichment Score (NES) | 1.6722245 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.32598528 |
| FWER p-Value | 0.253 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARID1A | 2630022 1690551 4810110 | 0 | 5.910 | 0.1886 | Yes | ||
| 2 | ILF3 | 940722 3190647 6520110 | 40 | 2.079 | 0.2528 | Yes | ||
| 3 | XRCC6 | 2850450 3870528 | 65 | 1.682 | 0.3051 | Yes | ||
| 4 | ATF4 | 730441 1740195 | 80 | 1.582 | 0.3549 | Yes | ||
| 5 | ATF7IP | 2690176 6770021 | 87 | 1.539 | 0.4036 | Yes | ||
| 6 | TNFRSF1A | 1090390 6520735 | 100 | 1.464 | 0.4497 | Yes | ||
| 7 | CTCF | 5340017 | 161 | 1.243 | 0.4861 | Yes | ||
| 8 | ELF1 | 4780450 | 249 | 1.032 | 0.5143 | Yes | ||
| 9 | SMARCC1 | 5080019 7100047 | 471 | 0.786 | 0.5275 | Yes | ||
| 10 | MED6 | 6840500 | 478 | 0.778 | 0.5520 | Yes | ||
| 11 | RUNX1 | 3840711 | 697 | 0.634 | 0.5605 | Yes | ||
| 12 | HNRPAB | 540504 | 740 | 0.612 | 0.5777 | Yes | ||
| 13 | CCNH | 3190347 | 764 | 0.603 | 0.5957 | Yes | ||
| 14 | MNAT1 | 730292 2350364 | 823 | 0.572 | 0.6108 | Yes | ||
| 15 | ARHGEF11 | 1780288 | 847 | 0.563 | 0.6276 | Yes | ||
| 16 | MAML1 | 2760008 | 876 | 0.553 | 0.6437 | Yes | ||
| 17 | MED12 | 4920095 | 903 | 0.542 | 0.6596 | Yes | ||
| 18 | MYO6 | 2190332 | 1416 | 0.355 | 0.6433 | Yes | ||
| 19 | ILF2 | 2900253 | 1452 | 0.347 | 0.6525 | Yes | ||
| 20 | TCF4 | 520021 | 1564 | 0.320 | 0.6567 | Yes | ||
| 21 | SMAD2 | 4200592 | 1581 | 0.312 | 0.6658 | Yes | ||
| 22 | RSF1 | 1580097 | 1591 | 0.309 | 0.6752 | Yes | ||
| 23 | SP1 | 6590017 | 1637 | 0.296 | 0.6822 | Yes | ||
| 24 | ARNTL | 3170463 | 1993 | 0.211 | 0.6697 | No | ||
| 25 | NSD1 | 4560519 4780672 | 2092 | 0.193 | 0.6706 | No | ||
| 26 | PPARGC1B | 3290114 | 2269 | 0.161 | 0.6662 | No | ||
| 27 | PHF5A | 2690519 | 2645 | 0.113 | 0.6496 | No | ||
| 28 | GATA4 | 1410215 2690609 | 3159 | 0.069 | 0.6241 | No | ||
| 29 | MRPL12 | 3120112 | 3303 | 0.060 | 0.6183 | No | ||
| 30 | BCL10 | 2360397 | 3328 | 0.059 | 0.6189 | No | ||
| 31 | CITED2 | 5670114 5130088 | 3359 | 0.057 | 0.6191 | No | ||
| 32 | NRIP1 | 3190039 | 3388 | 0.055 | 0.6193 | No | ||
| 33 | BPTF | 940541 4540152 | 4111 | 0.025 | 0.5812 | No | ||
| 34 | GLIS1 | 840471 | 4493 | 0.018 | 0.5612 | No | ||
| 35 | FOXD3 | 6550156 | 4790 | 0.014 | 0.5456 | No | ||
| 36 | SMARCA1 | 4230315 | 5194 | 0.010 | 0.5242 | No | ||
| 37 | RORB | 6400035 | 5304 | 0.009 | 0.5186 | No | ||
| 38 | ESRRG | 4010181 | 5662 | 0.007 | 0.4996 | No | ||
| 39 | PAX8 | 5050301 5290164 | 6032 | 0.006 | 0.4799 | No | ||
| 40 | FOXF2 | 1240091 | 6199 | 0.005 | 0.4711 | No | ||
| 41 | FOXE1 | 4810164 | 6444 | 0.004 | 0.4580 | No | ||
| 42 | MKL2 | 5700204 | 6731 | 0.003 | 0.4427 | No | ||
| 43 | TBX5 | 2900132 6370484 | 6908 | 0.003 | 0.4333 | No | ||
| 44 | GLI1 | 4560176 | 7168 | 0.002 | 0.4194 | No | ||
| 45 | BMP6 | 2100128 | 7649 | 0.001 | 0.3935 | No | ||
| 46 | RXRA | 3800471 | 8027 | -0.000 | 0.3732 | No | ||
| 47 | ELL3 | 1770484 | 8389 | -0.001 | 0.3537 | No | ||
| 48 | TRERF1 | 6370017 6940138 | 9032 | -0.003 | 0.3192 | No | ||
| 49 | NCOA6 | 1780333 6450110 | 9535 | -0.004 | 0.2922 | No | ||
| 50 | EHF | 4560088 | 9536 | -0.004 | 0.2923 | No | ||
| 51 | NPAS2 | 770670 | 9836 | -0.005 | 0.2763 | No | ||
| 52 | FOXH1 | 2060180 | 10511 | -0.007 | 0.2402 | No | ||
| 53 | CDK7 | 2640451 | 11027 | -0.009 | 0.2127 | No | ||
| 54 | NOTCH4 | 2450040 6370707 | 11342 | -0.010 | 0.1961 | No | ||
| 55 | HIF1A | 5670605 | 11601 | -0.012 | 0.1825 | No | ||
| 56 | THRAP3 | 6180309 | 11717 | -0.012 | 0.1767 | No | ||
| 57 | NUFIP1 | 6350241 | 12136 | -0.015 | 0.1546 | No | ||
| 58 | GLI2 | 3060632 | 12387 | -0.017 | 0.1417 | No | ||
| 59 | UTF1 | 5700593 | 12950 | -0.022 | 0.1121 | No | ||
| 60 | IL4 | 6020537 | 13283 | -0.026 | 0.0950 | No | ||
| 61 | FOXO3 | 2510484 4480451 | 13380 | -0.028 | 0.0907 | No | ||
| 62 | CREB5 | 2320368 | 13431 | -0.028 | 0.0889 | No | ||
| 63 | GTF2H1 | 2570746 2650148 | 14039 | -0.040 | 0.0574 | No | ||
| 64 | PLAGL1 | 3190082 6200193 | 14148 | -0.043 | 0.0529 | No | ||
| 65 | SMARCD3 | 4070497 | 14286 | -0.047 | 0.0470 | No | ||
| 66 | TCF3 | 5080022 | 14375 | -0.049 | 0.0438 | No | ||
| 67 | NARG1 | 5910563 6350095 | 14397 | -0.050 | 0.0443 | No | ||
| 68 | ERCC3 | 6900008 | 14917 | -0.068 | 0.0184 | No | ||
| 69 | DYRK1B | 5860706 | 15024 | -0.073 | 0.0150 | No | ||
| 70 | ELF4 | 4850193 | 15183 | -0.082 | 0.0091 | No | ||
| 71 | CLOCK | 1410070 2940292 3390075 | 15257 | -0.086 | 0.0079 | No | ||
| 72 | EPAS1 | 5290156 | 15387 | -0.094 | 0.0040 | No | ||
| 73 | HNF4A | 6620440 | 15585 | -0.111 | -0.0031 | No | ||
| 74 | EPC1 | 1570050 5290095 6900193 | 15826 | -0.135 | -0.0118 | No | ||
| 75 | TGFB1 | 1940162 | 15875 | -0.140 | -0.0099 | No | ||
| 76 | MAP2K3 | 5570193 | 15974 | -0.155 | -0.0102 | No | ||
| 77 | SPI1 | 1410397 | 15991 | -0.158 | -0.0061 | No | ||
| 78 | TRIM28 | 380292 | 16232 | -0.189 | -0.0130 | No | ||
| 79 | RBM14 | 5340731 5690315 | 16739 | -0.262 | -0.0319 | No | ||
| 80 | SUPT5H | 4150546 | 16987 | -0.306 | -0.0355 | No | ||
| 81 | SUPT4H1 | 6200056 | 17021 | -0.312 | -0.0273 | No | ||
| 82 | SQSTM1 | 6550056 | 17055 | -0.319 | -0.0189 | No | ||
| 83 | SCAP | 3800706 | 17258 | -0.361 | -0.0183 | No | ||
| 84 | SMAD3 | 6450671 | 17586 | -0.454 | -0.0215 | No | ||
| 85 | TP53 | 6130707 | 17702 | -0.490 | -0.0121 | No | ||
| 86 | SMARCD1 | 3060193 3850184 6400369 | 17805 | -0.536 | -0.0004 | No | ||
| 87 | ERCC2 | 2360750 4060390 6550138 | 17996 | -0.639 | 0.0097 | No | ||
| 88 | HMGA1 | 6580408 | 18109 | -0.743 | 0.0274 | No |

