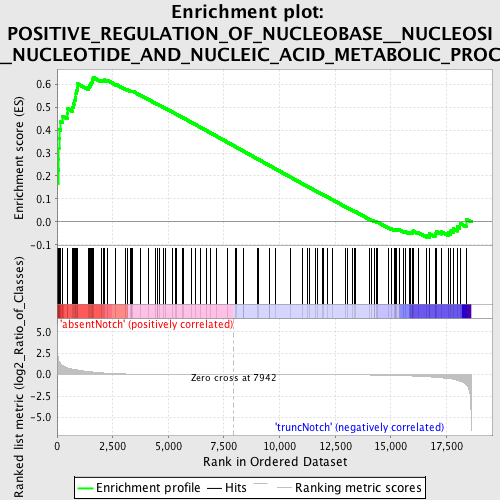
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | POSITIVE_REGULATION_OF_NUCLEOBASE__NUCLEOSIDE__NUCLEOTIDE_AND_NUCLEIC_ACID_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.62843627 |
| Normalized Enrichment Score (NES) | 1.5657476 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.33014148 |
| FWER p-Value | 0.94 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARID1A | 2630022 1690551 4810110 | 0 | 5.910 | 0.1692 | Yes | ||
| 2 | ILF3 | 940722 3190647 6520110 | 40 | 2.079 | 0.2266 | Yes | ||
| 3 | XRCC6 | 2850450 3870528 | 65 | 1.682 | 0.2735 | Yes | ||
| 4 | ATF4 | 730441 1740195 | 80 | 1.582 | 0.3181 | Yes | ||
| 5 | ATF7IP | 2690176 6770021 | 87 | 1.539 | 0.3618 | Yes | ||
| 6 | TNFRSF1A | 1090390 6520735 | 100 | 1.464 | 0.4031 | Yes | ||
| 7 | CTCF | 5340017 | 161 | 1.243 | 0.4354 | Yes | ||
| 8 | ELF1 | 4780450 | 249 | 1.032 | 0.4603 | Yes | ||
| 9 | SMARCC1 | 5080019 7100047 | 471 | 0.786 | 0.4708 | Yes | ||
| 10 | MED6 | 6840500 | 478 | 0.778 | 0.4928 | Yes | ||
| 11 | RUNX1 | 3840711 | 697 | 0.634 | 0.4992 | Yes | ||
| 12 | HNRPAB | 540504 | 740 | 0.612 | 0.5144 | Yes | ||
| 13 | CCNH | 3190347 | 764 | 0.603 | 0.5304 | Yes | ||
| 14 | MNAT1 | 730292 2350364 | 823 | 0.572 | 0.5437 | Yes | ||
| 15 | ARHGEF11 | 1780288 | 847 | 0.563 | 0.5586 | Yes | ||
| 16 | MAML1 | 2760008 | 876 | 0.553 | 0.5729 | Yes | ||
| 17 | MED12 | 4920095 | 903 | 0.542 | 0.5870 | Yes | ||
| 18 | CREBBP | 5690035 7040050 | 909 | 0.541 | 0.6022 | Yes | ||
| 19 | MYO6 | 2190332 | 1416 | 0.355 | 0.5850 | Yes | ||
| 20 | ILF2 | 2900253 | 1452 | 0.347 | 0.5931 | Yes | ||
| 21 | YAF2 | 940053 | 1490 | 0.338 | 0.6007 | Yes | ||
| 22 | TCF4 | 520021 | 1564 | 0.320 | 0.6060 | Yes | ||
| 23 | SMAD2 | 4200592 | 1581 | 0.312 | 0.6140 | Yes | ||
| 24 | RSF1 | 1580097 | 1591 | 0.309 | 0.6224 | Yes | ||
| 25 | SP1 | 6590017 | 1637 | 0.296 | 0.6284 | Yes | ||
| 26 | ARNTL | 3170463 | 1993 | 0.211 | 0.6153 | No | ||
| 27 | NSD1 | 4560519 4780672 | 2092 | 0.193 | 0.6155 | No | ||
| 28 | TNFSF13 | 2650170 | 2140 | 0.184 | 0.6182 | No | ||
| 29 | PPARGC1B | 3290114 | 2269 | 0.161 | 0.6159 | No | ||
| 30 | PHF5A | 2690519 | 2645 | 0.113 | 0.5989 | No | ||
| 31 | CAMKK2 | 5270047 | 3087 | 0.074 | 0.5772 | No | ||
| 32 | GATA4 | 1410215 2690609 | 3159 | 0.069 | 0.5753 | No | ||
| 33 | MRPL12 | 3120112 | 3303 | 0.060 | 0.5693 | No | ||
| 34 | BCL10 | 2360397 | 3328 | 0.059 | 0.5697 | No | ||
| 35 | CITED2 | 5670114 5130088 | 3359 | 0.057 | 0.5697 | No | ||
| 36 | NRIP1 | 3190039 | 3388 | 0.055 | 0.5698 | No | ||
| 37 | YWHAH | 1660133 2810053 | 3745 | 0.038 | 0.5516 | No | ||
| 38 | BPTF | 940541 4540152 | 4111 | 0.025 | 0.5326 | No | ||
| 39 | PPARGC1A | 4670040 | 4434 | 0.019 | 0.5158 | No | ||
| 40 | GLIS1 | 840471 | 4493 | 0.018 | 0.5131 | No | ||
| 41 | ACVR1 | 6840671 | 4525 | 0.017 | 0.5119 | No | ||
| 42 | FOXP3 | 940707 1850692 | 4608 | 0.016 | 0.5080 | No | ||
| 43 | FOXD3 | 6550156 | 4790 | 0.014 | 0.4986 | No | ||
| 44 | RGMB | 670504 4570450 | 4870 | 0.013 | 0.4947 | No | ||
| 45 | SMARCA1 | 4230315 | 5194 | 0.010 | 0.4775 | No | ||
| 46 | RORB | 6400035 | 5304 | 0.009 | 0.4719 | No | ||
| 47 | MYST4 | 1400563 2570687 3360458 6840402 | 5351 | 0.009 | 0.4697 | No | ||
| 48 | UBB | 4810138 | 5647 | 0.008 | 0.4539 | No | ||
| 49 | ESRRG | 4010181 | 5662 | 0.007 | 0.4534 | No | ||
| 50 | PAX8 | 5050301 5290164 | 6032 | 0.006 | 0.4336 | No | ||
| 51 | FOXF2 | 1240091 | 6199 | 0.005 | 0.4248 | No | ||
| 52 | FOXE1 | 4810164 | 6444 | 0.004 | 0.4117 | No | ||
| 53 | MKL2 | 5700204 | 6731 | 0.003 | 0.3964 | No | ||
| 54 | TBX5 | 2900132 6370484 | 6908 | 0.003 | 0.3869 | No | ||
| 55 | GLI1 | 4560176 | 7168 | 0.002 | 0.3730 | No | ||
| 56 | BMP6 | 2100128 | 7649 | 0.001 | 0.3471 | No | ||
| 57 | RXRA | 3800471 | 8027 | -0.000 | 0.3267 | No | ||
| 58 | ARNT2 | 1740594 | 8081 | -0.000 | 0.3238 | No | ||
| 59 | ELL3 | 1770484 | 8389 | -0.001 | 0.3073 | No | ||
| 60 | NFATC2 | 70450 540097 | 8399 | -0.001 | 0.3068 | No | ||
| 61 | ABCA1 | 6290156 | 9021 | -0.003 | 0.2733 | No | ||
| 62 | TRERF1 | 6370017 6940138 | 9032 | -0.003 | 0.2729 | No | ||
| 63 | NCOA6 | 1780333 6450110 | 9535 | -0.004 | 0.2459 | No | ||
| 64 | EHF | 4560088 | 9536 | -0.004 | 0.2460 | No | ||
| 65 | NPAS2 | 770670 | 9836 | -0.005 | 0.2300 | No | ||
| 66 | FOXH1 | 2060180 | 10511 | -0.007 | 0.1937 | No | ||
| 67 | CDK7 | 2640451 | 11027 | -0.009 | 0.1662 | No | ||
| 68 | EREG | 50519 4920129 | 11266 | -0.010 | 0.1536 | No | ||
| 69 | NOTCH4 | 2450040 6370707 | 11342 | -0.010 | 0.1498 | No | ||
| 70 | HIF1A | 5670605 | 11601 | -0.012 | 0.1362 | No | ||
| 71 | THRAP3 | 6180309 | 11717 | -0.012 | 0.1303 | No | ||
| 72 | ACVRL1 | 3290600 | 11913 | -0.013 | 0.1202 | No | ||
| 73 | RAD51 | 6110450 6980280 | 11989 | -0.014 | 0.1165 | No | ||
| 74 | NUFIP1 | 6350241 | 12136 | -0.015 | 0.1091 | No | ||
| 75 | GLI2 | 3060632 | 12387 | -0.017 | 0.0960 | No | ||
| 76 | UTF1 | 5700593 | 12950 | -0.022 | 0.0663 | No | ||
| 77 | IRF4 | 610133 | 13074 | -0.024 | 0.0603 | No | ||
| 78 | IL4 | 6020537 | 13283 | -0.026 | 0.0498 | No | ||
| 79 | FOXO3 | 2510484 4480451 | 13380 | -0.028 | 0.0454 | No | ||
| 80 | CREB5 | 2320368 | 13431 | -0.028 | 0.0436 | No | ||
| 81 | GTF2H1 | 2570746 2650148 | 14039 | -0.040 | 0.0119 | No | ||
| 82 | PLAGL1 | 3190082 6200193 | 14148 | -0.043 | 0.0073 | No | ||
| 83 | GLIS2 | 4150471 4810170 | 14250 | -0.046 | 0.0031 | No | ||
| 84 | SMARCD3 | 4070497 | 14286 | -0.047 | 0.0026 | No | ||
| 85 | TCF3 | 5080022 | 14375 | -0.049 | -0.0008 | No | ||
| 86 | NARG1 | 5910563 6350095 | 14397 | -0.050 | -0.0005 | No | ||
| 87 | ERCC3 | 6900008 | 14917 | -0.068 | -0.0266 | No | ||
| 88 | DYRK1B | 5860706 | 15024 | -0.073 | -0.0303 | No | ||
| 89 | ELF4 | 4850193 | 15183 | -0.082 | -0.0365 | No | ||
| 90 | NUP62 | 1240128 | 15205 | -0.083 | -0.0352 | No | ||
| 91 | UBE2N | 520369 2900047 | 15209 | -0.083 | -0.0330 | No | ||
| 92 | CLOCK | 1410070 2940292 3390075 | 15257 | -0.086 | -0.0331 | No | ||
| 93 | HIVEP3 | 3990551 | 15375 | -0.094 | -0.0367 | No | ||
| 94 | EPAS1 | 5290156 | 15387 | -0.094 | -0.0346 | No | ||
| 95 | HNF4A | 6620440 | 15585 | -0.111 | -0.0421 | No | ||
| 96 | BCL3 | 3990440 | 15669 | -0.119 | -0.0432 | No | ||
| 97 | EPC1 | 1570050 5290095 6900193 | 15826 | -0.135 | -0.0477 | No | ||
| 98 | TGFB1 | 1940162 | 15875 | -0.140 | -0.0463 | No | ||
| 99 | MAP2K3 | 5570193 | 15974 | -0.155 | -0.0472 | No | ||
| 100 | SPI1 | 1410397 | 15991 | -0.158 | -0.0435 | No | ||
| 101 | MYST1 | 2190397 | 15998 | -0.159 | -0.0393 | No | ||
| 102 | TRIM28 | 380292 | 16232 | -0.189 | -0.0465 | No | ||
| 103 | MYST3 | 5270500 | 16625 | -0.244 | -0.0607 | No | ||
| 104 | BMP7 | 870039 | 16732 | -0.261 | -0.0590 | No | ||
| 105 | RBM14 | 5340731 5690315 | 16739 | -0.262 | -0.0518 | No | ||
| 106 | SUPT5H | 4150546 | 16987 | -0.306 | -0.0564 | No | ||
| 107 | SUPT4H1 | 6200056 | 17021 | -0.312 | -0.0492 | No | ||
| 108 | SQSTM1 | 6550056 | 17055 | -0.319 | -0.0418 | No | ||
| 109 | SCAP | 3800706 | 17258 | -0.361 | -0.0424 | No | ||
| 110 | SMAD3 | 6450671 | 17586 | -0.454 | -0.0471 | No | ||
| 111 | TP53 | 6130707 | 17702 | -0.490 | -0.0393 | No | ||
| 112 | SMARCD1 | 3060193 3850184 6400369 | 17805 | -0.536 | -0.0294 | No | ||
| 113 | ERCC2 | 2360750 4060390 6550138 | 17996 | -0.639 | -0.0214 | No | ||
| 114 | HMGA1 | 6580408 | 18109 | -0.743 | -0.0062 | No | ||
| 115 | EGR1 | 4610347 | 18402 | -1.171 | 0.0116 | No |

