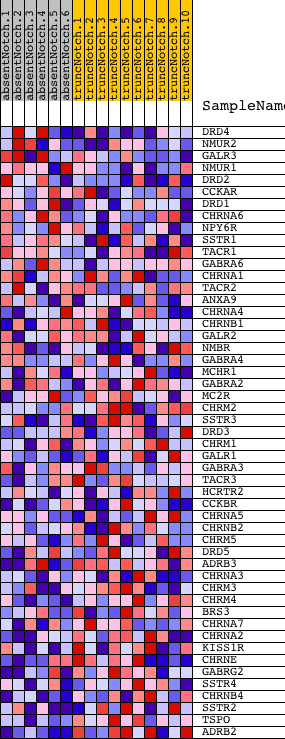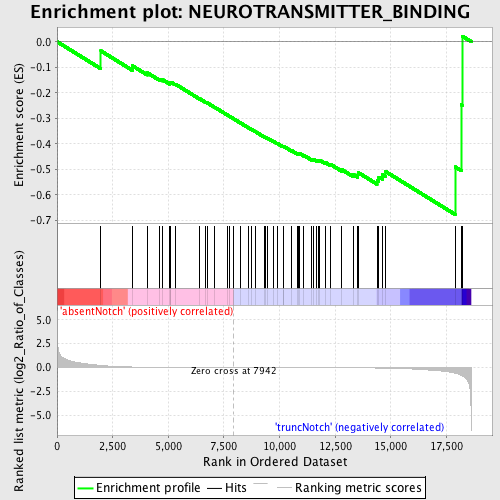
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | truncNotch |
| GeneSet | NEUROTRANSMITTER_BINDING |
| Enrichment Score (ES) | -0.67693794 |
| Normalized Enrichment Score (NES) | -1.4710102 |
| Nominal p-value | 0.02244898 |
| FDR q-value | 0.9892104 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DRD4 | 6350390 | 1968 | 0.218 | -0.0359 | No | ||
| 2 | NMUR2 | 1580717 | 3382 | 0.056 | -0.0942 | No | ||
| 3 | GALR3 | 4850113 | 4057 | 0.027 | -0.1218 | No | ||
| 4 | NMUR1 | 1500403 | 4610 | 0.016 | -0.1464 | No | ||
| 5 | DRD2 | 5890369 | 4750 | 0.014 | -0.1493 | No | ||
| 6 | CCKAR | 4210079 | 5064 | 0.011 | -0.1626 | No | ||
| 7 | DRD1 | 430025 | 5090 | 0.011 | -0.1605 | No | ||
| 8 | CHRNA6 | 5340092 | 5117 | 0.011 | -0.1584 | No | ||
| 9 | NPY6R | 4560136 | 5321 | 0.009 | -0.1664 | No | ||
| 10 | SSTR1 | 2630471 | 6414 | 0.004 | -0.2238 | No | ||
| 11 | TACR1 | 70358 3840411 | 6667 | 0.004 | -0.2363 | No | ||
| 12 | GABRA6 | 4120520 | 6751 | 0.003 | -0.2397 | No | ||
| 13 | CHRNA1 | 1170025 3610364 5220292 | 7069 | 0.002 | -0.2560 | No | ||
| 14 | TACR2 | 1740358 | 7671 | 0.001 | -0.2882 | No | ||
| 15 | ANXA9 | 1190195 | 7742 | 0.000 | -0.2918 | No | ||
| 16 | CHRNA4 | 730075 2680091 | 7913 | 0.000 | -0.3009 | No | ||
| 17 | CHRNB1 | 1190671 | 7942 | -0.000 | -0.3024 | No | ||
| 18 | GALR2 | 6450739 | 8251 | -0.001 | -0.3188 | No | ||
| 19 | NMBR | 6180315 | 8601 | -0.002 | -0.3370 | No | ||
| 20 | GABRA4 | 6290465 | 8756 | -0.002 | -0.3447 | No | ||
| 21 | MCHR1 | 1780022 | 8924 | -0.002 | -0.3529 | No | ||
| 22 | GABRA2 | 3290167 | 9338 | -0.004 | -0.3740 | No | ||
| 23 | MC2R | 2970504 | 9388 | -0.004 | -0.3755 | No | ||
| 24 | CHRM2 | 870750 | 9448 | -0.004 | -0.3774 | No | ||
| 25 | SSTR3 | 5420064 | 9714 | -0.005 | -0.3902 | No | ||
| 26 | DRD3 | 4780402 | 9893 | -0.005 | -0.3981 | No | ||
| 27 | CHRM1 | 4280619 | 10158 | -0.006 | -0.4104 | No | ||
| 28 | GALR1 | 6020452 | 10191 | -0.006 | -0.4102 | No | ||
| 29 | GABRA3 | 4810142 | 10548 | -0.007 | -0.4270 | No | ||
| 30 | TACR3 | 4780017 | 10811 | -0.008 | -0.4385 | No | ||
| 31 | HCRTR2 | 2350463 | 10833 | -0.008 | -0.4370 | No | ||
| 32 | CCKBR | 2760128 | 10913 | -0.009 | -0.4385 | No | ||
| 33 | CHRNA5 | 580195 | 11092 | -0.009 | -0.4451 | No | ||
| 34 | CHRNB2 | 580204 3120739 | 11453 | -0.011 | -0.4610 | No | ||
| 35 | CHRM5 | 5390333 | 11514 | -0.011 | -0.4606 | No | ||
| 36 | DRD5 | 2970133 | 11677 | -0.012 | -0.4655 | No | ||
| 37 | ADRB3 | 6900072 | 11728 | -0.012 | -0.4642 | No | ||
| 38 | CHRNA3 | 6760100 | 11795 | -0.013 | -0.4637 | No | ||
| 39 | CHRM3 | 6110725 | 12074 | -0.014 | -0.4740 | No | ||
| 40 | CHRM4 | 1770427 | 12281 | -0.016 | -0.4800 | No | ||
| 41 | BRS3 | 1740403 | 12795 | -0.021 | -0.5010 | No | ||
| 42 | CHRNA7 | 2970446 | 13311 | -0.027 | -0.5202 | No | ||
| 43 | CHRNA2 | 5050315 | 13523 | -0.030 | -0.5218 | No | ||
| 44 | KISS1R | 2480601 | 13528 | -0.030 | -0.5122 | No | ||
| 45 | CHRNE | 3190170 | 14380 | -0.049 | -0.5423 | Yes | ||
| 46 | GABRG2 | 2350402 4210204 6130279 6550037 | 14460 | -0.051 | -0.5301 | Yes | ||
| 47 | SSTR4 | 6100398 | 14624 | -0.056 | -0.5208 | Yes | ||
| 48 | CHRNB4 | 6370110 | 14772 | -0.062 | -0.5089 | Yes | ||
| 49 | SSTR2 | 4590687 | 17892 | -0.581 | -0.4905 | Yes | ||
| 50 | TSPO | 110692 3390452 | 18167 | -0.806 | -0.2464 | Yes | ||
| 51 | ADRB2 | 3290373 | 18208 | -0.843 | 0.0220 | Yes |